Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000903A_C01 KMC000903A_c01
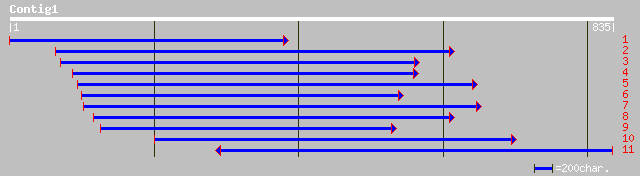
(831 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_197752.1| cation-transporting ATPase; protein id: At5g236... 186 5e-46
gb|AAF65410.1|AF238314_1 putative cation-transporting ATPase Cta... 62 1e-08
gb|EAA29171.1| hypothetical protein [Neurospora crassa] 61 2e-08
ref|NP_010883.1| Sensitivity to a killer toxin (SMK toxin) produ... 60 3e-08
pir||T19187 hypothetical protein C10C6.6 - Caenorhabditis elegans 60 5e-08
>ref|NP_197752.1| cation-transporting ATPase; protein id: At5g23630.1 [Arabidopsis
thaliana] gi|12229714|sp|Q9LT02|ATX1_ARATH Potential
cation-transporting ATPase gi|8809697|dbj|BAA97238.1|
cation-transporting ATPase [Arabidopsis thaliana]
Length = 1179
Score = 186 bits (471), Expect = 5e-46
Identities = 87/114 (76%), Positives = 98/114 (85%)
Frame = -3
Query: 805 VNMMLQVATFAVNYMGHPFNQSIPENKPFLYALLAAVVFFTAIASDIFRDLNDWLKLVPL 626
V+MMLQVATFAVNYMGHPFNQSI ENKPF YAL+A FFT IASD+FRDLND LKLVPL
Sbjct: 1066 VSMMLQVATFAVNYMGHPFNQSIRENKPFFYALIAGAGFFTVIASDLFRDLNDSLKLVPL 1125
Query: 625 PAGLRNKLLTWAFLMFLACFSWERLLRWAFPGKIPAWKKRQRVAVSNLEKKKQL 464
P GLR+KLL WA LMF+ C+SWERLLRWAFPGKI +WK +QR +NLEKKK++
Sbjct: 1126 PQGLRDKLLIWASLMFIICYSWERLLRWAFPGKISSWKHKQRAVTANLEKKKKV 1179
>gb|AAF65410.1|AF238314_1 putative cation-transporting ATPase CtaA [Dictyostelium discoideum]
Length = 1208
Score = 61.6 bits (148), Expect = 1e-08
Identities = 35/64 (54%), Positives = 40/64 (61%)
Frame = -3
Query: 814 FLHVNMMLQVATFAVNYMGHPFNQSIPENKPFLYALLAAVVFFTAIASDIFRDLNDWLKL 635
FL N M QVATFAVNY GHPF QS+ ENKP LYAL +A++I LN L+L
Sbjct: 1114 FLMSNAM-QVATFAVNYKGHPFMQSLSENKPLLYALSFVWGLGLLLATEIIPPLNSMLEL 1172
Query: 634 VPLP 623
V P
Sbjct: 1173 VEFP 1176
>gb|EAA29171.1| hypothetical protein [Neurospora crassa]
Length = 1318
Score = 61.2 bits (147), Expect = 2e-08
Identities = 33/123 (26%), Positives = 66/123 (52%), Gaps = 3/123 (2%)
Frame = -3
Query: 805 VNMMLQVATFAVNYMGHPFNQSIPENKPFLYALLAAVVFFTAIASDIFRDLNDWLKLVPL 626
+ ++ Q++TFAVNY G PF +S+ ENK Y ++ A ++++ +LN+ +KLVP
Sbjct: 1165 LQLIQQISTFAVNYQGRPFRESLSENKGMFYGIVGVTAIAFACSTEMVPELNEAMKLVPF 1224
Query: 625 PAGLRNKLLTWAFLMFLACFSWERLLRWAF---PGKIPAWKKRQRVAVSNLEKKKQL*VV 455
+ + L ++AC+ E + ++ F + A ++ ++ L K ++L +
Sbjct: 1225 KEEFKWTMTIVMILDYVACWLIEVVFKYLFSDLKARDIAVRREDQLERERLRKAEELKKL 1284
Query: 454 EEM 446
EE+
Sbjct: 1285 EEV 1287
>ref|NP_010883.1| Sensitivity to a killer toxin (SMK toxin) produced by Pichia
Farinosa; Spf1p [Saccharomyces cerevisiae]
gi|731415|sp|P39986|ATC6_YEAST Probable
cation-transporting ATPase 1 gi|1077608|pir||S50428
probable Ca2+-transporting ATPase (EC 3.6.3.8) SPF1 -
yeast (Saccharomyces cerevisiae)
gi|602398|gb|AAB64508.1| P-type ATPase; YEL031W
[Saccharomyces cerevisiae]
Length = 1215
Score = 60.5 bits (145), Expect = 3e-08
Identities = 33/107 (30%), Positives = 54/107 (49%)
Frame = -3
Query: 805 VNMMLQVATFAVNYMGHPFNQSIPENKPFLYALLAAVVFFTAIASDIFRDLNDWLKLVPL 626
+ ++ QV+TFAVNY G PF ++I NK Y LL A A++ +LN+ +K VP+
Sbjct: 1108 IQLVQQVSTFAVNYQGEPFRENIRSNKGMYYGLLGVTGLALASATEFLPELNEAMKFVPM 1167
Query: 625 PAGLRNKLLTWAFLMFLACFSWERLLRWAFPGKIPAWKKRQRVAVSN 485
+ KL L F + E ++ F P+ Q+V +++
Sbjct: 1168 TDDFKIKLTLTLLLDFFGSWGVEHFFKFFFMDDKPSDISVQQVKIAS 1214
>pir||T19187 hypothetical protein C10C6.6 - Caenorhabditis elegans
Length = 1157
Score = 59.7 bits (143), Expect = 5e-08
Identities = 35/88 (39%), Positives = 49/88 (54%)
Frame = -3
Query: 805 VNMMLQVATFAVNYMGHPFNQSIPENKPFLYALLAAVVFFTAIASDIFRDLNDWLKLVPL 626
++M LQV TFAVNY G PF +S+ ENK LY+++ + +AS DL +LV L
Sbjct: 1064 ISMALQVCTFAVNYRGRPFMESLFENKAMLYSIMFSGGAVFTLASGQATDLMIQFELVVL 1123
Query: 625 PAGLRNKLLTWAFLMFLACFSWERLLRW 542
P LRN LL + C+ +R L +
Sbjct: 1124 PEALRNALLMCVTADLVICYIIDRGLNF 1151
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 675,313,187
Number of Sequences: 1393205
Number of extensions: 14228327
Number of successful extensions: 37129
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 35657
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37082
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 42922608498
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)