Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000901A_C01 KMC000901A_c01
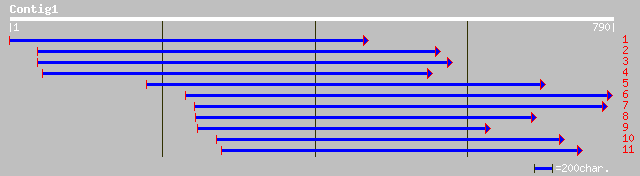
(783 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_197599.1| MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN; protei... 181 1e-44
gb|AAA97413.1| molybdenum cofactor biosynthesis enzyme gi|446912... 179 5e-44
gb|AAF73075.1|AF268595_1 molybdenum cofactor biosynthesis protei... 172 4e-42
pdb|1O8N|A Chain A, The Active Site Of The Molybdenum Cofactor B... 134 2e-30
pdb|1O8Q|A Chain A, The Active Site Of The Molybdenum Cofactor B... 126 3e-28
>ref|NP_197599.1| MOLYBDOPTERIN BIOSYNTHESIS CNX1 PROTEIN; protein id: At5g20990.1,
supported by cDNA: gi_1263313 [Arabidopsis thaliana]
gi|26454618|sp|Q39054|CNX1_ARATH Molybdopterin
biosynthesis CNX1 protein (Molybdenum cofactor
biosynthesis enzyme CNX1)
Length = 670
Score = 181 bits (459), Expect = 1e-44
Identities = 90/116 (77%), Positives = 100/116 (85%), Gaps = 4/116 (3%)
Frame = -1
Query: 783 ATKELIEKETPALLHVMMQESLKVTPSAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 604
ATK++IE+ETP LL VMMQESLK+TP AMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL
Sbjct: 553 ATKKVIERETPGLLFVMMQESLKITPFAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 612
Query: 603 LPALKHGLKQLRGDKREKHPRHVPHAE-TVPTDAWEQSYMSASGGG---SEVSCSC 448
LPALKH LKQ++GDKREKHP+H+PHAE T+PTD W+QSY SA G E CSC
Sbjct: 613 LPALKHALKQIKGDKREKHPKHIPHAEATLPTDTWDQSYKSAYETGEKKEEAGCSC 668
>gb|AAA97413.1| molybdenum cofactor biosynthesis enzyme gi|4469123|emb|CAB38312.1|
molybdenum cofactor biosynthesis enzyme [Arabidopsis
thaliana]
Length = 670
Score = 179 bits (453), Expect = 5e-44
Identities = 89/116 (76%), Positives = 99/116 (84%), Gaps = 4/116 (3%)
Frame = -1
Query: 783 ATKELIEKETPALLHVMMQESLKVTPSAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 604
ATK++IE+ETP LL VMMQESLK+TP AMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL
Sbjct: 553 ATKKVIERETPGLLFVMMQESLKITPFAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 612
Query: 603 LPALKHGLKQLRGDKREKHPRHVPHAE-TVPTDAWEQSYMSASGGG---SEVSCSC 448
LPALKH LKQ++GDKREKHP+H+PHAE T+PTD W+QSY A G E CSC
Sbjct: 613 LPALKHALKQIKGDKREKHPKHIPHAEATLPTDTWDQSYKLAYETGEKKEEAGCSC 668
>gb|AAF73075.1|AF268595_1 molybdenum cofactor biosynthesis protein Cnx1 [Hordeum vulgare]
Length = 659
Score = 172 bits (437), Expect = 4e-42
Identities = 85/112 (75%), Positives = 94/112 (83%)
Frame = -1
Query: 783 ATKELIEKETPALLHVMMQESLKVTPSAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 604
ATK +IEKE P L VM+QESLKVTP AMLSR+A GIRGSTLIINMPGNPNAVAECMEAL
Sbjct: 549 ATKCVIEKEAPGLAFVMIQESLKVTPFAMLSRAACGIRGSTLIINMPGNPNAVAECMEAL 608
Query: 603 LPALKHGLKQLRGDKREKHPRHVPHAETVPTDAWEQSYMSASGGGSEVSCSC 448
LPALKH +KQL+GDKREK+PRHVPHAE P D WE+S+ +AS GG CSC
Sbjct: 609 LPALKHAMKQLKGDKREKNPRHVPHAEAAPVDQWERSFRAASSGG---GCSC 657
>pdb|1O8N|A Chain A, The Active Site Of The Molybdenum Cofactor Biosynthetic
Protein Domain Cnx1g gi|28948326|pdb|1O8N|B Chain B, The
Active Site Of The Molybdenum Cofactor Biosynthetic
Protein Domain Cnx1g gi|28948327|pdb|1O8N|C Chain C, The
Active Site Of The Molybdenum Cofactor Biosynthetic
Protein Domain Cnx1g
Length = 167
Score = 134 bits (336), Expect = 2e-30
Identities = 66/76 (86%), Positives = 72/76 (93%)
Frame = -1
Query: 783 ATKELIEKETPALLHVMMQESLKVTPSAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 604
ATK++IE+ETP LL VMMQESLK+TP AMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL
Sbjct: 92 ATKKVIERETPGLLFVMMQESLKITPFAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 151
Query: 603 LPALKHGLKQLRGDKR 556
LPALKH LKQ++GDKR
Sbjct: 152 LPALKHALKQIKGDKR 167
>pdb|1O8Q|A Chain A, The Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g gi|28948329|pdb|1O8Q|B Chain B, The
Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g gi|28948330|pdb|1O8Q|C Chain C, The
Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g gi|28948331|pdb|1O8Q|D Chain D, The
Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g gi|28948332|pdb|1O8Q|E Chain E, The
Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g gi|28948333|pdb|1O8Q|F Chain F, The
Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g gi|28948334|pdb|1O8Q|G Chain G, The
Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g gi|28948335|pdb|1O8Q|H Chain H, The
Active Site Of The Molybdenum Cofactor Biosenthetic
Protein Domain Cnx1g
Length = 160
Score = 126 bits (317), Expect = 3e-28
Identities = 62/73 (84%), Positives = 69/73 (93%)
Frame = -1
Query: 783 ATKELIEKETPALLHVMMQESLKVTPSAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 604
ATK++IE+ETP LL VMMQE+LK+TP AMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL
Sbjct: 88 ATKKVIERETPGLLFVMMQEALKITPFAMLSRSAAGIRGSTLIINMPGNPNAVAECMEAL 147
Query: 603 LPALKHGLKQLRG 565
LPALKH LKQ++G
Sbjct: 148 LPALKHALKQIKG 160
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 651,598,168
Number of Sequences: 1393205
Number of extensions: 14365769
Number of successful extensions: 50549
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 47975
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 50528
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38935490438
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)