Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
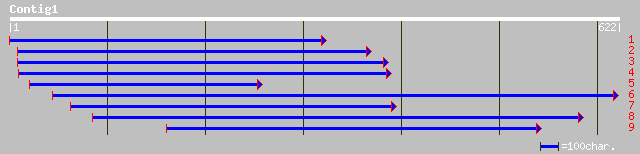
Query= KMC000897A_C01 KMC000897A_c01
(605 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA75572.1| putative start codon [Medicago truncatula] 145 5e-34
gb|AAO22748.1| unknown protein [Arabidopsis thaliana] 106 3e-22
ref|NP_198846.1| putative protein; protein id: At5g40300.1 [Arab... 103 2e-21
ref|NP_201088.1| putative protein; protein id: At5g62820.1 [Arab... 96 4e-19
ref|NP_181174.1| hypothetical protein; protein id: At2g36330.1 [... 59 6e-08
>emb|CAA75572.1| putative start codon [Medicago truncatula]
Length = 234
Score = 145 bits (365), Expect = 5e-34
Identities = 69/113 (61%), Positives = 91/113 (80%)
Frame = -2
Query: 604 YDYKEFKYSLSVNVIGFVYSGLQICDLVKYLITKIHTLNLRIRGYFSVAMDQGFAYLLMS 425
Y + EF+YS + NVIGFVYSGLQIC LV YLITK HT+N +++GYF+VA+DQ AY+LMS
Sbjct: 118 YKWDEFRYSFAANVIGFVYSGLQICHLVMYLITKKHTINPKLQGYFNVAIDQTLAYILMS 177
Query: 424 ASSSAASTIHISRSYWVGEGVHKFTDMASASIALSFLAFLAFASASIVSGLML 266
ASSSAA+ H+ + YW+ G F +MA+AS+++SFLAF AFA AS+VSG++L
Sbjct: 178 ASSSAATAAHLLKDYWLEHGADTFIEMANASVSMSFLAFGAFALASLVSGIIL 230
>gb|AAO22748.1| unknown protein [Arabidopsis thaliana]
Length = 283
Score = 106 bits (264), Expect = 3e-22
Identities = 56/111 (50%), Positives = 77/111 (68%)
Frame = -2
Query: 598 YKEFKYSLSVNVIGFVYSGLQICDLVKYLITKIHTLNLRIRGYFSVAMDQGFAYLLMSAS 419
YKE+++ LSVNV+ FVYS Q CDL +L+ + H ++ +R F +DQ AYLLMSAS
Sbjct: 170 YKEYRFCLSVNVVAFVYSSFQACDLAYHLVKEKHLISHHLRPLFEFIIDQVLAYLLMSAS 229
Query: 418 SSAASTIHISRSYWVGEGVHKFTDMASASIALSFLAFLAFASASIVSGLML 266
++A + + S W G +FT+MASASIA+SFLAFLAFA +S++SG L
Sbjct: 230 TAAVTRVDDWVSNW---GKDEFTEMASASIAMSFLAFLAFAFSSLISGYNL 277
>ref|NP_198846.1| putative protein; protein id: At5g40300.1 [Arabidopsis thaliana]
gi|10178139|dbj|BAB11584.1| gene_id:MPO12.1~unknown
protein [Arabidopsis thaliana]
Length = 270
Score = 103 bits (257), Expect = 2e-21
Identities = 55/113 (48%), Positives = 73/113 (63%)
Frame = -2
Query: 604 YDYKEFKYSLSVNVIGFVYSGLQICDLVKYLITKIHTLNLRIRGYFSVAMDQGFAYLLMS 425
Y+YKEF++ L+ NVIGFVYSG ICDLV L T I +R + +DQ AYLL S
Sbjct: 155 YNYKEFRFCLAANVIGFVYSGFMICDLVYLLSTSIRRSRHNLRHFLEFGLDQMLAYLLAS 214
Query: 424 ASSSAASTIHISRSYWVGEGVHKFTDMASASIALSFLAFLAFASASIVSGLML 266
AS+SA+ + +S W G KF D+A AS+ALS+++F+AFA S+ SG L
Sbjct: 215 ASTSASIRVDDWQSNW---GADKFPDLARASVALSYVSFVAFAFCSLASGYAL 264
>ref|NP_201088.1| putative protein; protein id: At5g62820.1 [Arabidopsis thaliana]
Length = 297
Score = 95.9 bits (237), Expect = 4e-19
Identities = 55/114 (48%), Positives = 73/114 (63%), Gaps = 1/114 (0%)
Frame = -2
Query: 604 YD-YKEFKYSLSVNVIGFVYSGLQICDLVKYLITKIHTLNLRIRGYFSVAMDQGFAYLLM 428
YD YKE++Y L+VNVI FVYS + CD Y+ + + +N F +MDQ AYLLM
Sbjct: 181 YDRYKEYRYCLAVNVIAFVYSAFEACDAACYIAKESYMINCGFHDLFVFSMDQLLAYLLM 240
Query: 427 SASSSAASTIHISRSYWVGEGVHKFTDMASASIALSFLAFLAFASASIVSGLML 266
SASS AA+ + S W G +FT MA+ASIA+SFLAF AFA ++++S L
Sbjct: 241 SASSCAATRVDDWVSNW---GKDEFTQMATASIAVSFLAFGAFAVSALISSYRL 291
>ref|NP_181174.1| hypothetical protein; protein id: At2g36330.1 [Arabidopsis
thaliana] gi|25408462|pir||D84779 hypothetical protein
At2g36330 [imported] - Arabidopsis thaliana
gi|4510344|gb|AAD21433.1| hypothetical protein
[Arabidopsis thaliana]
Length = 431
Score = 58.5 bits (140), Expect = 6e-08
Identities = 36/91 (39%), Positives = 53/91 (57%)
Frame = -2
Query: 538 QICDLVKYLITKIHTLNLRIRGYFSVAMDQGFAYLLMSASSSAASTIHISRSYWVGEGVH 359
Q CDL +L+ + H ++ +R F +DQ ++A + + S W G
Sbjct: 348 QACDLAYHLVKEKHLISHHLRPLFEFIIDQ----------ATAVTRVDDWVSNW---GKD 394
Query: 358 KFTDMASASIALSFLAFLAFASASIVSGLML 266
+FT+MASASIA+SFLAFLAFA +S++SG L
Sbjct: 395 EFTEMASASIAMSFLAFLAFAFSSLISGYNL 425
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 506,109,155
Number of Sequences: 1393205
Number of extensions: 10695760
Number of successful extensions: 28217
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 27469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28197
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)