Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000889A_C01 KMC000889A_c01
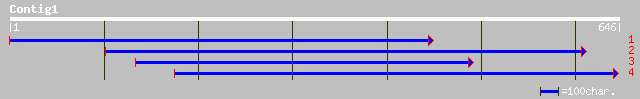
(646 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA92739.1| hypothetical protein [Oryza sativa (japonica cul... 140 1e-32
dbj|BAC22206.1| hypothetical protein~predicted by FGENESH etc. [... 131 9e-30
gb|EAA10144.1| agCP15288 [Anopheles gambiae str. PEST] 44 0.002
gb|AAC32373.1| centriole associated protein CEP110 [Homo sapiens] 42 0.007
ref|NP_008949.2| centrosomal protein 1; centriole associated pro... 42 0.007
>dbj|BAA92739.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 445
Score = 140 bits (354), Expect = 1e-32
Identities = 70/82 (85%), Positives = 78/82 (94%)
Frame = -1
Query: 646 RKMAEAKERNSTPSVPSDMQAVLKRCENLEKEVRSLKLNLSFMNRKDSEQTKQIEDLQKQ 467
RKMAEAKER+STP +PSD+Q VLKRCE LEKEVRSLKLNLSFMNRKDSEQTKQIE+LQKQ
Sbjct: 362 RKMAEAKERSSTPVIPSDIQVVLKRCETLEKEVRSLKLNLSFMNRKDSEQTKQIEELQKQ 421
Query: 466 NEELADEKERLLEEIERILSET 401
NE+L +EKERLLEEIERI+S+T
Sbjct: 422 NEDLVEEKERLLEEIERIVSDT 443
>dbj|BAC22206.1| hypothetical protein~predicted by FGENESH etc. [Oryza sativa
(japonica cultivar-group)]
Length = 524
Score = 131 bits (329), Expect = 9e-30
Identities = 67/81 (82%), Positives = 76/81 (93%)
Frame = -1
Query: 643 KMAEAKERNSTPSVPSDMQAVLKRCENLEKEVRSLKLNLSFMNRKDSEQTKQIEDLQKQN 464
+MAEAKER+STP +PSD+Q VLKRCE LEKEVRSLKLNLSFMN KDSEQTKQIE+LQKQN
Sbjct: 443 RMAEAKERSSTPVIPSDIQVVLKRCETLEKEVRSLKLNLSFMN-KDSEQTKQIEELQKQN 501
Query: 463 EELADEKERLLEEIERILSET 401
E+L +EKERLLEEIERI+S+T
Sbjct: 502 EDLVEEKERLLEEIERIVSDT 522
>gb|EAA10144.1| agCP15288 [Anopheles gambiae str. PEST]
Length = 1842
Score = 43.5 bits (101), Expect = 0.002
Identities = 24/59 (40%), Positives = 38/59 (63%)
Frame = -1
Query: 568 ENLEKEVRSLKLNLSFMNRKDSEQTKQIEDLQKQNEELADEKERLLEEIERILSETEKM 392
E+LE+E+ +LK + S +DSE +K ++DL+KQNEEL E + + E +L + E M
Sbjct: 1071 ESLEREMAALK-SKSATGAEDSESSKILQDLKKQNEELTTEIHQQSRKFEELLQKHETM 1128
>gb|AAC32373.1| centriole associated protein CEP110 [Homo sapiens]
Length = 994
Score = 42.0 bits (97), Expect = 0.007
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Frame = -1
Query: 610 PSVPSDMQAVLKRCENLEKEVRSLKLNLSF-MNRKDSEQTKQIEDLQKQNEELADE--KE 440
P +P+D++A+L+R ENLE E+ SLK NL F MN E+ + +E E +E
Sbjct: 849 PELPADLEAILERNENLEGELESLKENLPFTMNEGPFEEKLNFSQVHIMDEHWRGEALRE 908
Query: 439 RLLEEIERILSE 404
+L +R+ ++
Sbjct: 909 KLRHREDRLKAQ 920
>ref|NP_008949.2| centrosomal protein 1; centriole associated protein [Homo sapiens]
Length = 994
Score = 42.0 bits (97), Expect = 0.007
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Frame = -1
Query: 610 PSVPSDMQAVLKRCENLEKEVRSLKLNLSF-MNRKDSEQTKQIEDLQKQNEELADE--KE 440
P +P+D++A+L+R ENLE E+ SLK NL F MN E+ + +E E +E
Sbjct: 849 PELPADLEAILERNENLEGELESLKENLPFTMNEGPFEEKLNFSQVHIMDEHWRGEALRE 908
Query: 439 RLLEEIERILSE 404
+L +R+ ++
Sbjct: 909 KLRHREDRLKAQ 920
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 473,443,297
Number of Sequences: 1393205
Number of extensions: 9038878
Number of successful extensions: 37163
Number of sequences better than 10.0: 559
Number of HSP's better than 10.0 without gapping: 31784
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36784
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27291941472
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)