Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000888A_C01 KMC000888A_c01
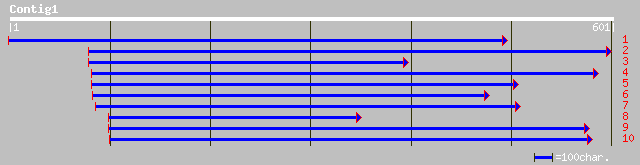
(601 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL91244.1| unknown protein [Arabidopsis thaliana] 180 1e-44
ref|NP_180358.1| unknown protein; protein id: At2g27910.1 [Arabi... 180 1e-44
gb|AAK20049.1|AC025783_9 unknown protein [Oryza sativa (japonica... 179 2e-44
dbj|BAB24670.1| unnamed protein product [Mus musculus] 76 3e-13
dbj|BAC36038.1| unnamed protein product [Mus musculus] 76 3e-13
>gb|AAL91244.1| unknown protein [Arabidopsis thaliana]
Length = 1124
Score = 180 bits (457), Expect = 1e-44
Identities = 84/98 (85%), Positives = 93/98 (94%)
Frame = -3
Query: 599 EGLSRVKRCSDEGRALMSLDLQVLINGLHHFVSLNVKPKLQMVETFIKAYYLPETEYVHW 420
EGLSR+KRC+DEGR LMSLDLQVLINGL HFV VKPKLQ+VETFIKAYYLPETEYVHW
Sbjct: 1023 EGLSRIKRCTDEGRVLMSLDLQVLINGLQHFVPTKVKPKLQIVETFIKAYYLPETEYVHW 1082
Query: 419 ARAHPEYSKSQVVGLINLVATMKGWKRKTRLDILEKIE 306
ARAHPEY+K+QVVGL+NLVATMKGWKRKTRL+++EKIE
Sbjct: 1083 ARAHPEYTKAQVVGLVNLVATMKGWKRKTRLEVIEKIE 1120
>ref|NP_180358.1| unknown protein; protein id: At2g27910.1 [Arabidopsis thaliana]
gi|25370737|pir||D84678 hypothetical protein At2g27910
[imported] - Arabidopsis thaliana
gi|4510425|gb|AAD21511.1| unknown protein [Arabidopsis
thaliana]
Length = 140
Score = 180 bits (457), Expect = 1e-44
Identities = 84/98 (85%), Positives = 93/98 (94%)
Frame = -3
Query: 599 EGLSRVKRCSDEGRALMSLDLQVLINGLHHFVSLNVKPKLQMVETFIKAYYLPETEYVHW 420
EGLSR+KRC+DEGR LMSLDLQVLINGL HFV VKPKLQ+VETFIKAYYLPETEYVHW
Sbjct: 39 EGLSRIKRCTDEGRVLMSLDLQVLINGLQHFVPTKVKPKLQIVETFIKAYYLPETEYVHW 98
Query: 419 ARAHPEYSKSQVVGLINLVATMKGWKRKTRLDILEKIE 306
ARAHPEY+K+QVVGL+NLVATMKGWKRKTRL+++EKIE
Sbjct: 99 ARAHPEYTKAQVVGLVNLVATMKGWKRKTRLEVIEKIE 136
>gb|AAK20049.1|AC025783_9 unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 336
Score = 179 bits (454), Expect = 2e-44
Identities = 85/98 (86%), Positives = 94/98 (95%)
Frame = -3
Query: 599 EGLSRVKRCSDEGRALMSLDLQVLINGLHHFVSLNVKPKLQMVETFIKAYYLPETEYVHW 420
EGLSRVKRC+DEGRALMSLDLQVLINGL H VS NV+PKLQ+V+TFIKAYYLPETEYVHW
Sbjct: 236 EGLSRVKRCTDEGRALMSLDLQVLINGLLHIVSANVRPKLQIVDTFIKAYYLPETEYVHW 295
Query: 419 ARAHPEYSKSQVVGLINLVATMKGWKRKTRLDILEKIE 306
AR+HPEYSKSQVVGL+NLVATMKGWKRKTRL+ +E+IE
Sbjct: 296 ARSHPEYSKSQVVGLVNLVATMKGWKRKTRLETIERIE 333
>dbj|BAB24670.1| unnamed protein product [Mus musculus]
Length = 343
Score = 76.3 bits (186), Expect = 3e-13
Identities = 40/98 (40%), Positives = 54/98 (54%)
Frame = -3
Query: 599 EGLSRVKRCSDEGRALMSLDLQVLINGLHHFVSLNVKPKLQMVETFIKAYYLPETEYVHW 420
EG + VK+CS+EGRALM LD Q + L + P + VET+IKAYYL E + W
Sbjct: 240 EGYANVKKCSNEGRALMQLDFQQFLMKLEKLTDIRPIPDKEFVETYIKAYYLTENDMERW 299
Query: 419 ARAHPEYSKSQVVGLINLVATMKGWKRKTRLDILEKIE 306
+ H EYS Q+ L+N V +K R +L I+
Sbjct: 300 IKEHREYSTKQLTNLVN-VCLGSHINKKARQKLLAAID 336
>dbj|BAC36038.1| unnamed protein product [Mus musculus]
Length = 964
Score = 76.3 bits (186), Expect = 3e-13
Identities = 40/98 (40%), Positives = 54/98 (54%)
Frame = -3
Query: 599 EGLSRVKRCSDEGRALMSLDLQVLINGLHHFVSLNVKPKLQMVETFIKAYYLPETEYVHW 420
EG + VK+CS+EGRALM LD Q + L + P + VET+IKAYYL E + W
Sbjct: 861 EGYANVKKCSNEGRALMQLDFQQFLMKLEKLTDIRPIPDKEFVETYIKAYYLTENDMERW 920
Query: 419 ARAHPEYSKSQVVGLINLVATMKGWKRKTRLDILEKIE 306
+ H EYS Q+ L+N V +K R +L I+
Sbjct: 921 IKEHREYSTKQLTNLVN-VCLGSHINKKARQKLLAAID 957
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 533,233,542
Number of Sequences: 1393205
Number of extensions: 11574029
Number of successful extensions: 27847
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 27253
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27839
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)