Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000877A_C01 KMC000877A_c01
(797 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
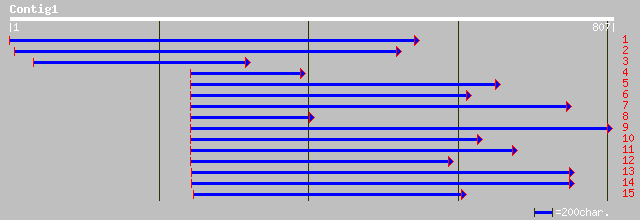
ref|NP_179257.1| nodulin-like protein; protein id: At2g16660.1, ... 220 2e-56
ref|NP_195221.1| putative protein; protein id: At4g34950.1 [Arab... 220 2e-56
gb|AAN62343.1|AF506028_10 nodulin-like protein [Poncirus trifoli... 135 7e-31
ref|NP_177616.1| hypothetical protein; protein id: At1g74780.1 [... 122 5e-27
ref|NP_173328.1| hypothetical protein; protein id: At1g18940.1 [... 119 5e-26
>ref|NP_179257.1| nodulin-like protein; protein id: At2g16660.1, supported by cDNA:
gi_17979282, supported by cDNA: gi_20465249 [Arabidopsis
thaliana] gi|25370604|pir||F84542 nodulin-like protein
[imported] - Arabidopsis thaliana
gi|4581109|gb|AAD24599.1| nodulin-like protein
[Arabidopsis thaliana] gi|17979283|gb|AAL49867.1|
putative nodulin protein [Arabidopsis thaliana]
gi|20465250|gb|AAM19945.1| At2g16660/T24I21.7
[Arabidopsis thaliana] gi|23463081|gb|AAN33210.1|
At2g16660/T24I21.7 [Arabidopsis thaliana]
Length = 546
Score = 220 bits (560), Expect = 2e-56
Identities = 107/140 (76%), Positives = 123/140 (87%)
Frame = -1
Query: 797 MAVGYILLAMALPGSLYIGSIVVGICYGVRLAITVPTASELFGLKYYGLIYNILILNLPL 618
MAVGYIL+A+A+P SLYIGS+VVG+CYGVRLAITVPTASELFGLKYYGLIYNIL+LNLPL
Sbjct: 407 MAVGYILMALAVPNSLYIGSMVVGVCYGVRLAITVPTASELFGLKYYGLIYNILVLNLPL 466
Query: 617 GSFLFSGLLAGILYDREATATEGGGNTCVGGHCYRLVFIVMAAACIVGFFLDILLSIRTQ 438
GSFLFSGLLAG LYD EAT T GGGNTCVG HCYRL+FIVMA A ++G LD++L+ RT+
Sbjct: 467 GSFLFSGLLAGFLYDAEATPTPGGGNTCVGAHCYRLIFIVMALASVIGVGLDLVLAYRTK 526
Query: 437 NVYNKISMSKKSQKSLGTSS 378
+Y KI SKKS+KS G+ S
Sbjct: 527 EIYAKIHASKKSKKSGGSLS 546
>ref|NP_195221.1| putative protein; protein id: At4g34950.1 [Arabidopsis thaliana]
gi|7486862|pir||T10241 hypothetical protein T11I11.190 -
Arabidopsis thaliana gi|5123712|emb|CAB45456.1| putative
protein [Arabidopsis thaliana]
gi|7270446|emb|CAB80212.1| putative protein [Arabidopsis
thaliana]
Length = 567
Score = 220 bits (560), Expect = 2e-56
Identities = 107/137 (78%), Positives = 123/137 (89%)
Frame = -1
Query: 797 MAVGYILLAMALPGSLYIGSIVVGICYGVRLAITVPTASELFGLKYYGLIYNILILNLPL 618
MAVGY+L+A+ALPGSLYIGS+VVG+CYGVRLAITVPTASELFGLKYYGLIYNILILN+PL
Sbjct: 428 MAVGYLLMALALPGSLYIGSMVVGVCYGVRLAITVPTASELFGLKYYGLIYNILILNMPL 487
Query: 617 GSFLFSGLLAGILYDREATATEGGGNTCVGGHCYRLVFIVMAAACIVGFFLDILLSIRTQ 438
GSFLFSGLLAG+LYD EAT T GGGNTCVG HC+R+VFIVMA A I+G LD+LL+ RT+
Sbjct: 488 GSFLFSGLLAGLLYDAEATPTPGGGNTCVGAHCFRIVFIVMAFASIIGVGLDLLLAYRTK 547
Query: 437 NVYNKISMSKKSQKSLG 387
+Y KI SKK++KS G
Sbjct: 548 GIYAKIHASKKTKKSGG 564
>gb|AAN62343.1|AF506028_10 nodulin-like protein [Poncirus trifoliata]
Length = 564
Score = 135 bits (340), Expect = 7e-31
Identities = 65/126 (51%), Positives = 92/126 (72%)
Frame = -1
Query: 797 MAVGYILLAMALPGSLYIGSIVVGICYGVRLAITVPTASELFGLKYYGLIYNILILNLPL 618
M + ++L A AL G+LY +I++G+C GV ++ VPTASELFGLK++GLIYN ++L P+
Sbjct: 427 MILTFLLYASALSGTLYAATILLGVCCGVIYSLMVPTASELFGLKHFGLIYNFILLGNPI 486
Query: 617 GSFLFSGLLAGILYDREATATEGGGNTCVGGHCYRLVFIVMAAACIVGFFLDILLSIRTQ 438
G+ LFSGLLAG LYD E AT+ G +TC+G C+RL F+V+A C +G L I+L+IR +
Sbjct: 487 GALLFSGLLAGKLYDAE--ATKQGSSTCIGAECFRLTFLVLAGVCGLGTILSIILTIRIR 544
Query: 437 NVYNKI 420
VY +
Sbjct: 545 PVYQML 550
>ref|NP_177616.1| hypothetical protein; protein id: At1g74780.1 [Arabidopsis
thaliana] gi|25370605|pir||B96777 hypothetical protein
F25A4.25 [imported] - Arabidopsis thaliana
gi|5882744|gb|AAD55297.1|AC008263_28 Strong similarity
to gb|AF031243 nodule-specific protein (Nlj70) from
Lotus japonicus and is a member of the PF|00083 Sugar
(and other) transporter family. EST gb|Z37715 comes
from this gene. [Arabidopsis thaliana]
Length = 533
Score = 122 bits (307), Expect = 5e-27
Identities = 53/130 (40%), Positives = 89/130 (67%)
Frame = -1
Query: 797 MAVGYILLAMALPGSLYIGSIVVGICYGVRLAITVPTASELFGLKYYGLIYNILILNLPL 618
M++G++++A G+LY+GS++VG+CYG + ++ SELFG+++ G I+N + + P+
Sbjct: 403 MSIGHLIIASGFQGNLYVGSVIVGVCYGSQWSLMPTITSELFGIRHMGTIFNTISVASPI 462
Query: 617 GSFLFSGLLAGILYDREATATEGGGNTCVGGHCYRLVFIVMAAACIVGFFLDILLSIRTQ 438
GS++FS L G +YD+ A+ G GNTC G HC+RL FI+MA+ GF + I+L RT+
Sbjct: 463 GSYIFSVRLIGYIYDKTAS---GEGNTCYGSHCFRLSFIIMASVAFFGFLVAIVLFFRTK 519
Query: 437 NVYNKISMSK 408
+Y +I + +
Sbjct: 520 TLYRQILVKR 529
>ref|NP_173328.1| hypothetical protein; protein id: At1g18940.1 [Arabidopsis
thaliana] gi|8778273|gb|AAF79282.1|AC068602_5 F14D16.8
[Arabidopsis thaliana]
Length = 526
Score = 119 bits (298), Expect = 5e-26
Identities = 54/126 (42%), Positives = 83/126 (65%)
Frame = -1
Query: 797 MAVGYILLAMALPGSLYIGSIVVGICYGVRLAITVPTASELFGLKYYGLIYNILILNLPL 618
M +G++++A G+LY GSI+VGICYG + ++ SELFG+K+ G IYN + + P+
Sbjct: 396 MTIGHLIIASGFQGNLYPGSIIVGICYGSQWSLMPTITSELFGVKHMGTIYNTISIASPM 455
Query: 617 GSFLFSGLLAGILYDREATATEGGGNTCVGGHCYRLVFIVMAAACIVGFFLDILLSIRTQ 438
GS++FS L G +YDR G GNTC G HC+RL ++V+A+ +GF + +L RT+
Sbjct: 456 GSYIFSVRLIGYIYDRTII---GEGNTCYGPHCFRLAYVVIASVAFLGFLVSCVLVFRTK 512
Query: 437 NVYNKI 420
+Y +I
Sbjct: 513 TIYRQI 518
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 735,020,330
Number of Sequences: 1393205
Number of extensions: 17252805
Number of successful extensions: 47036
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 44182
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46899
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40336047648
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)