Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000873A_C01 KMC000873A_c01
(703 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
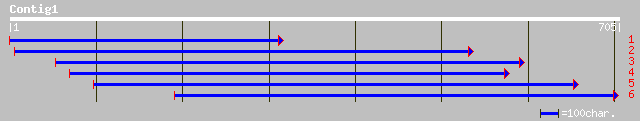
gb|AAF64489.1| prespore protein MF12 [Dictyostelium discoideum] 40 0.024
emb|CAB05388.1| FCA gamma [Arabidopsis thaliana] 39 0.070
ref|NP_680711.1| Flowering time control protein (FCA); protein i... 39 0.070
emb|CAB05395.1| FCA alpha 2 [Arabidopsis thaliana] 39 0.070
ref|NP_193363.2| Flowering time control protein (FCA); protein i... 39 0.070
>gb|AAF64489.1| prespore protein MF12 [Dictyostelium discoideum]
Length = 1287
Score = 40.4 bits (93), Expect = 0.024
Identities = 32/96 (33%), Positives = 40/96 (41%)
Frame = -3
Query: 695 QQSRTQSQPSVLAAQQVAQIQQAQPQSNLQGQVFNQQQMQLPSVSSSFQAYGVTGHQNNQ 516
QQ + Q Q +L QQ+ Q QQ Q Q LQ Q QQQ QL S++ Q NN
Sbjct: 728 QQQQQQQQQQILQQQQLQQ-QQQQQQQQLQQQQLQQQQQQLQQQSTNKQKPSQEFPPNNS 786
Query: 515 EVGFKQLQASAISGGDPRRYSQFQGMNTAQELMWKN 408
KQ Q ++ Q Q Q+ KN
Sbjct: 787 AKQKKQQQQQQQQQQQQQQQQQQQQQQRQQQQHIKN 822
>emb|CAB05388.1| FCA gamma [Arabidopsis thaliana]
Length = 747
Score = 38.9 bits (89), Expect = 0.070
Identities = 33/117 (28%), Positives = 43/117 (36%), Gaps = 20/117 (17%)
Frame = -3
Query: 695 QQSRTQSQPSVLAAQQVAQIQQAQPQSNLQGQVFNQQQMQLPSVSSSFQAYGVTGH---- 528
QQ + Q Q Q Q+Q Q Q Q + QQ+Q P SS + G + +
Sbjct: 632 QQKQQQHQEKPTIQQSQTQLQPLQQQPQQVQQQYQGQQLQQPFYSSLYPTPGASHNTQYP 691
Query: 527 ----------------QNNQEVGFKQLQASAISGGDPRRYSQFQGMNTAQELMWKNK 405
QN Q+ + A S D R Q + QELMWKNK
Sbjct: 692 SLPVGQNSQFPMSGIGQNAQDYARTHIPVGAASMNDISRTQ--QSRQSPQELMWKNK 746
>ref|NP_680711.1| Flowering time control protein (FCA); protein id: At4g16280.2
[Arabidopsis thaliana] gi|7430376|pir||E71429 probable
FCA gamma - Arabidopsis thaliana
gi|2244986|emb|CAB10407.1| FCA gamma protein
[Arabidopsis thaliana] gi|7268377|emb|CAB78670.1| FCA
gamma protein [Arabidopsis thaliana]
Length = 747
Score = 38.9 bits (89), Expect = 0.070
Identities = 33/117 (28%), Positives = 43/117 (36%), Gaps = 20/117 (17%)
Frame = -3
Query: 695 QQSRTQSQPSVLAAQQVAQIQQAQPQSNLQGQVFNQQQMQLPSVSSSFQAYGVTGH---- 528
QQ + Q Q Q Q+Q Q Q Q + QQ+Q P SS + G + +
Sbjct: 632 QQKQQQHQEKPTIQQSQTQLQPLQQQPQQVQQQYQGQQLQQPFYSSLYPTPGASHNTQYP 691
Query: 527 ----------------QNNQEVGFKQLQASAISGGDPRRYSQFQGMNTAQELMWKNK 405
QN Q+ + A S D R Q + QELMWKNK
Sbjct: 692 SLPVGQNSQFPMSGIGQNAQDYARTHIPVGAASMNDISRTQ--QSRQSPQELMWKNK 746
>emb|CAB05395.1| FCA alpha 2 [Arabidopsis thaliana]
Length = 505
Score = 38.9 bits (89), Expect = 0.070
Identities = 33/117 (28%), Positives = 43/117 (36%), Gaps = 20/117 (17%)
Frame = -3
Query: 695 QQSRTQSQPSVLAAQQVAQIQQAQPQSNLQGQVFNQQQMQLPSVSSSFQAYGVTGH---- 528
QQ + Q Q Q Q+Q Q Q Q + QQ+Q P SS + G + +
Sbjct: 390 QQKQQQHQEKPTIQQSQTQLQPLQQQPQQVQQQYQGQQLQQPFYSSLYPTPGASHNTQYP 449
Query: 527 ----------------QNNQEVGFKQLQASAISGGDPRRYSQFQGMNTAQELMWKNK 405
QN Q+ + A S D R Q + QELMWKNK
Sbjct: 450 SLPVGQNSQFPMSGIGQNAQDYARTHIPVGAASMNDISRTQ--QSRQSPQELMWKNK 504
>ref|NP_193363.2| Flowering time control protein (FCA); protein id: At4g16280.1
[Arabidopsis thaliana]
Length = 505
Score = 38.9 bits (89), Expect = 0.070
Identities = 33/117 (28%), Positives = 43/117 (36%), Gaps = 20/117 (17%)
Frame = -3
Query: 695 QQSRTQSQPSVLAAQQVAQIQQAQPQSNLQGQVFNQQQMQLPSVSSSFQAYGVTGH---- 528
QQ + Q Q Q Q+Q Q Q Q + QQ+Q P SS + G + +
Sbjct: 390 QQKQQQHQEKPTIQQSQTQLQPLQQQPQQVQQQYQGQQLQQPFYSSLYPTPGASHNTQYP 449
Query: 527 ----------------QNNQEVGFKQLQASAISGGDPRRYSQFQGMNTAQELMWKNK 405
QN Q+ + A S D R Q + QELMWKNK
Sbjct: 450 SLPVGQNSQFPMSGIGQNAQDYARTHIPVGAASMNDISRTQ--QSRQSPQELMWKNK 504
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 610,493,662
Number of Sequences: 1393205
Number of extensions: 13197565
Number of successful extensions: 32584
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 28629
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31633
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 32250355128
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)