Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
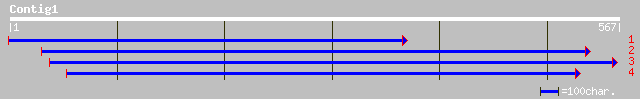
Query= KMC000870A_C01 KMC000870A_c01
(565 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568378.1| putative protein; protein id: At5g19620.1 [Arab... 243 2e-63
ref|NP_488933.1| hypothetical protein [Nostoc sp. PCC 7120] gi|2... 160 1e-38
gb|ZP_00106444.1| hypothetical protein [Nostoc punctiforme] 153 2e-36
ref|ZP_00072147.1| hypothetical protein [Trichodesmium erythraeu... 151 5e-36
ref|NP_190002.1| putative protein; protein id: At3g44160.1 [Arab... 147 7e-35
>ref|NP_568378.1| putative protein; protein id: At5g19620.1 [Arabidopsis thaliana]
gi|13430586|gb|AAK25915.1|AF360205_1 unknown protein
[Arabidopsis thaliana] gi|14532858|gb|AAK64111.1| unknown
protein [Arabidopsis thaliana]
Length = 732
Score = 243 bits (619), Expect = 2e-63
Identities = 115/139 (82%), Positives = 124/139 (88%)
Frame = -1
Query: 529 RARKGVEIGPARILLSLSGGHVKGNFSPYEAFAIGGTNSVRGYEEGGVGSGRSYVVGSGE 350
RARKG+ IGPAR L SLSGGHV G FSP+EAF IGGTNSVRGYEEG VGSGRSYVVGSGE
Sbjct: 594 RARKGIHIGPARFLFSLSGGHVVGKFSPHEAFVIGGTNSVRGYEEGAVGSGRSYVVGSGE 653
Query: 349 ISFPMFGPVEGVVFSDYGTDLGSGPTVPGDPAGARGKPGSGHGYGFGIRVDSPLGPLRLE 170
+SFP+ GPVEGV+F+DYGTD+GSG TVPGDPAGAR KPGSG+GYG G+RVDSPLGPLRLE
Sbjct: 654 LSFPVRGPVEGVIFTDYGTDMGSGSTVPGDPAGARLKPGSGYGYGLGVRVDSPLGPLRLE 713
Query: 169 YAFNDQKDKRFHFGVGHRN 113
YAFNDQ RFHFGVG RN
Sbjct: 714 YAFNDQHAGRFHFGVGLRN 732
>ref|NP_488933.1| hypothetical protein [Nostoc sp. PCC 7120] gi|25532565|pir||AE2417
hypothetical protein alr4893 [imported] - Nostoc sp.
(strain PCC 7120) gi|17134030|dbj|BAB76592.1|
ORF_ID:alr4893~hypothetical protein [Nostoc sp. PCC
7120]
Length = 676
Score = 160 bits (405), Expect = 1e-38
Identities = 76/129 (58%), Positives = 92/129 (70%)
Frame = -1
Query: 502 PARILLSLSGGHVKGNFSPYEAFAIGGTNSVRGYEEGGVGSGRSYVVGSGEISFPMFGPV 323
P L++ G V GN PYE F +GG+NSVRGY+ G VGSGRSYV+ S E FP+ V
Sbjct: 547 PQVFALNVQAGTVLGNLPPYETFNLGGSNSVRGYDAGNVGSGRSYVLASAEYRFPIVPIV 606
Query: 322 EGVVFSDYGTDLGSGPTVPGDPAGARGKPGSGHGYGFGIRVDSPLGPLRLEYAFNDQKDK 143
GV+F+D+ +DLGSG TV G+PAG RGKPGSG GYG GIRVDSPLG +R +Y NDQ +
Sbjct: 607 GGVLFADFASDLGSGDTVLGNPAGVRGKPGSGFGYGAGIRVDSPLGLIRADYGINDQGES 666
Query: 142 RFHFGVGHR 116
R H G+G R
Sbjct: 667 RVHLGIGQR 675
>gb|ZP_00106444.1| hypothetical protein [Nostoc punctiforme]
Length = 688
Score = 153 bits (386), Expect = 2e-36
Identities = 71/129 (55%), Positives = 91/129 (70%)
Frame = -1
Query: 502 PARILLSLSGGHVKGNFSPYEAFAIGGTNSVRGYEEGGVGSGRSYVVGSGEISFPMFGPV 323
P ++L G V GN PYE+F +GG+NSVRGY+ G VGSGRSYV+ S E FP+ V
Sbjct: 559 PQVFAVNLQAGTVIGNLPPYESFNLGGSNSVRGYDSGDVGSGRSYVLASAEYRFPVLPIV 618
Query: 322 EGVVFSDYGTDLGSGPTVPGDPAGARGKPGSGHGYGFGIRVDSPLGPLRLEYAFNDQKDK 143
GV+F+D+ +DLGSG TV GDPAG R KPG G GYG G+R++SPLG +R +Y NDQ +
Sbjct: 619 GGVLFADFASDLGSGDTVLGDPAGVRDKPGYGFGYGAGVRLNSPLGLIRADYGINDQGES 678
Query: 142 RFHFGVGHR 116
+ H G+G R
Sbjct: 679 KVHLGIGQR 687
>ref|ZP_00072147.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 932
Score = 151 bits (382), Expect = 5e-36
Identities = 68/130 (52%), Positives = 93/130 (71%)
Frame = -1
Query: 505 GPARILLSLSGGHVKGNFSPYEAFAIGGTNSVRGYEEGGVGSGRSYVVGSGEISFPMFGP 326
GP + ++ GH+ G+ PYEAFA+GGTN+VRGY+EG V +GR++V+G+ E FP+F
Sbjct: 802 GPQALAFNIQAGHIIGDLPPYEAFALGGTNTVRGYDEGSVAAGRTFVLGTVEYRFPVFKF 861
Query: 325 VEGVVFSDYGTDLGSGPTVPGDPAGARGKPGSGHGYGFGIRVDSPLGPLRLEYAFNDQKD 146
+ G +F D T S +V G+P G R KPG G GYG G+RV+SPLGP+R++YA ND+ D
Sbjct: 862 LGGALFVDAATVFDSQRSVIGNPGGVREKPGDGIGYGGGLRVNSPLGPIRIDYAINDEGD 921
Query: 145 KRFHFGVGHR 116
RFHFG+G R
Sbjct: 922 TRFHFGIGER 931
>ref|NP_190002.1| putative protein; protein id: At3g44160.1 [Arabidopsis thaliana]
gi|11357663|pir||T49132 hypothetical protein F26G5.110 -
Arabidopsis thaliana gi|7635461|emb|CAB88424.1| putative
protein [Arabidopsis thaliana]
Length = 435
Score = 147 bits (372), Expect = 7e-35
Identities = 70/134 (52%), Positives = 94/134 (69%)
Frame = -1
Query: 526 ARKGVEIGPARILLSLSGGHVKGNFSPYEAFAIGGTNSVRGYEEGGVGSGRSYVVGSGEI 347
A KGV GPA +L SL+GG + G+ +PY+AFAIGG SVRGY EG VGSGRS +V + E+
Sbjct: 263 ASKGVRFGPAFLLASLTGGSIVGDMAPYQAFAIGGLGSVRGYGEGAVGSGRSCLVANTEL 322
Query: 346 SFPMFGPVEGVVFSDYGTDLGSGPTVPGDPAGARGKPGSGHGYGFGIRVDSPLGPLRLEY 167
+ P+ EG +F D GTDLGS VPG+P+ +GKPG G+G+G+G+R SPLG L+++Y
Sbjct: 323 ALPLNKMTEGTIFLDCGTDLGSSRLVPGNPSMRQGKPGFGYGFGYGLRFKSPLGHLQVDY 382
Query: 166 AFNDQKDKRFHFGV 125
A N K +F +
Sbjct: 383 AINAFNQKTLYFDI 396
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 549,406,724
Number of Sequences: 1393205
Number of extensions: 14602601
Number of successful extensions: 88382
Number of sequences better than 10.0: 1289
Number of HSP's better than 10.0 without gapping: 58470
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 80103
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)