Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000850A_C01 KMC000850A_c01
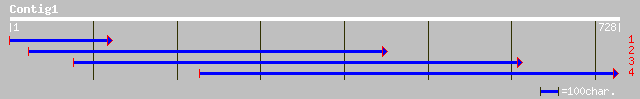
(725 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL49957.1| GTP cyclohydrolase I [Lycopersicon esculentum] 197 2e-49
ref|NP_187383.1| GTP cyclohydrolase I; protein id: At3g07270.1, ... 192 5e-48
gb|AAM03126.1|AF489530_1 GTP cyclohydrolase I [Arabidopsis thali... 192 5e-48
ref|NP_301283.1| putative GTP cyclohydrolase I [Mycobacterium le... 100 2e-20
emb|CAC86189.1| GTP cyclohydrolase I [Danio rerio] 100 4e-20
>gb|AAL49957.1| GTP cyclohydrolase I [Lycopersicon esculentum]
Length = 456
Score = 197 bits (500), Expect = 2e-49
Identities = 102/162 (62%), Positives = 118/162 (71%), Gaps = 17/162 (10%)
Frame = -3
Query: 702 FFVDSVNVNKEVSFNG-----------------EKIYSEVNLPFWSQCEHHLLPFHGVVH 574
+F++ N N E+ NG + I SE+NL FWSQCEHHLLPF GVVH
Sbjct: 293 WFMNFRNSNLEMKLNGFVRSRIDTRSPQGGNFNDGICSELNLSFWSQCEHHLLPFQGVVH 352
Query: 573 IGYFISDGFKPITKSLLQSMVHFYGFKLQVQERLTRQIAETVSPLLGGDVIVVVEASHTC 394
IGY SDG P+ + L+QS+VHFYGFKLQVQER+TRQIAETVS LG D+IVVVEA+HTC
Sbjct: 353 IGYHSSDGVNPVGRPLVQSVVHFYGFKLQVQERVTRQIAETVSSFLGEDIIVVVEANHTC 412
Query: 393 MISRGIEKFGSSTATIAVLGRFSTDIAARASFLQGIPSPTSS 268
MISRGIEKFGS+TAT AVLGRFSTD ARA FLQ +P S+
Sbjct: 413 MISRGIEKFGSNTATFAVLGRFSTDPVARAKFLQSLPDSGSA 454
Score = 34.7 bits (78), Expect = 1.4
Identities = 22/79 (27%), Positives = 38/79 (47%), Gaps = 1/79 (1%)
Frame = -3
Query: 633 NLPFWSQCEHHLLPFHGVVHIGYFISDGFKPITKSLLQSMVHFYGFKLQVQERLTRQIAE 454
+L +S CE LLPF H+GY + G + + S L + + +LQ +RL ++
Sbjct: 106 DLDLFSYCESCLLPFQVKCHVGY-VPSGKRVVGLSKLSRVADIFAKRLQSPQRLADEVCT 164
Query: 453 TVS-PLLGGDVIVVVEASH 400
+ + V VV++ H
Sbjct: 165 ALQHGIKPTGVAVVLQCMH 183
>ref|NP_187383.1| GTP cyclohydrolase I; protein id: At3g07270.1, supported by cDNA:
gi_19909131, supported by cDNA: gi_20466831 [Arabidopsis
thaliana] gi|6642638|gb|AAF20219.1|AC012395_6 GTP
cyclohydrolase I [Arabidopsis thaliana]
gi|20466832|gb|AAM20733.1| GTP cyclohydrolase I
[Arabidopsis thaliana]
Length = 466
Score = 192 bits (487), Expect = 5e-48
Identities = 95/134 (70%), Positives = 114/134 (84%), Gaps = 2/134 (1%)
Frame = -3
Query: 681 VNKEVSFNGEKIYSEVNLPFWSQCEHHLLPFHGVVHIGYFISDGFKP--ITKSLLQSMVH 508
VN EV ++++ E+N+PFWS CEHHLLPF+GVVHIGYF ++G P + SL++++VH
Sbjct: 321 VNGEVK--EKRLHCELNMPFWSMCEHHLLPFYGVVHIGYFCAEGSNPNPVGSSLMKAIVH 378
Query: 507 FYGFKLQVQERLTRQIAETVSPLLGGDVIVVVEASHTCMISRGIEKFGSSTATIAVLGRF 328
FYGFKLQVQER+TRQIAET+SPL+GGDVIVV EA HTCMISRGIEKFGSSTATIAVLGRF
Sbjct: 379 FYGFKLQVQERMTRQIAETLSPLVGGDVIVVAEAGHTCMISRGIEKFGSSTATIAVLGRF 438
Query: 327 STDIAARASFLQGI 286
S+D +ARA FL I
Sbjct: 439 SSDNSARAMFLDKI 452
Score = 39.3 bits (90), Expect = 0.057
Identities = 28/94 (29%), Positives = 43/94 (44%), Gaps = 1/94 (1%)
Frame = -3
Query: 633 NLPFWSQCEHHLLPFHGVVHIGYFISDGFKPITKSLLQSMVHFYGFKLQVQERLTRQIAE 454
+L +S CE LLPFH HIGY + G + + S + + +LQ +RL I
Sbjct: 109 DLDHYSYCESCLLPFHVKCHIGY-VPSGQRVLGLSKFSRVTDVFAKRLQDPQRLADDICS 167
Query: 453 TVSP-LLGGDVIVVVEASHTCMISRGIEKFGSST 355
+ + V VV+E SH S ++ S+
Sbjct: 168 ALQHWVKPAGVAVVLECSHIHFPSLDLDSLNLSS 201
>gb|AAM03126.1|AF489530_1 GTP cyclohydrolase I [Arabidopsis thaliana]
Length = 466
Score = 192 bits (487), Expect = 5e-48
Identities = 95/134 (70%), Positives = 114/134 (84%), Gaps = 2/134 (1%)
Frame = -3
Query: 681 VNKEVSFNGEKIYSEVNLPFWSQCEHHLLPFHGVVHIGYFISDGFKP--ITKSLLQSMVH 508
VN EV ++++ E+N+PFWS CEHHLLPF+GVVHIGYF ++G P + SL++++VH
Sbjct: 321 VNGEVK--EKRLHCELNMPFWSMCEHHLLPFYGVVHIGYFCAEGSNPNPVGSSLMKAIVH 378
Query: 507 FYGFKLQVQERLTRQIAETVSPLLGGDVIVVVEASHTCMISRGIEKFGSSTATIAVLGRF 328
FYGFKLQVQER+TRQIAET+SPL+GGDVIVV EA HTCMISRGIEKFGSSTATIAVLGRF
Sbjct: 379 FYGFKLQVQERMTRQIAETLSPLVGGDVIVVAEAGHTCMISRGIEKFGSSTATIAVLGRF 438
Query: 327 STDIAARASFLQGI 286
S+D +ARA FL I
Sbjct: 439 SSDNSARAMFLDKI 452
Score = 39.3 bits (90), Expect = 0.057
Identities = 28/94 (29%), Positives = 43/94 (44%), Gaps = 1/94 (1%)
Frame = -3
Query: 633 NLPFWSQCEHHLLPFHGVVHIGYFISDGFKPITKSLLQSMVHFYGFKLQVQERLTRQIAE 454
+L +S CE LLPFH HIGY + G + + S + + +LQ +RL I
Sbjct: 109 DLDHYSYCESCLLPFHVKCHIGY-VPSGQRVLGLSKFSRVTDVFAKRLQDPQRLADDICS 167
Query: 453 TVSP-LLGGDVIVVVEASHTCMISRGIEKFGSST 355
+ + V VV+E SH S ++ S+
Sbjct: 168 ALQHWVKPAGVAVVLECSHIHFPSLDLDSLNLSS 201
>ref|NP_301283.1| putative GTP cyclohydrolase I [Mycobacterium leprae]
gi|6016112|sp|O69531|GCH1_MYCLE GTP cyclohydrolase I
(GTP-CH-I) gi|25290287|pir||G86936 probable GTP
cyclohydrolase I [imported] - Mycobacterium leprae
gi|3097224|emb|CAA18795.1| GTP cyclohydrolase I
[Mycobacterium leprae] gi|13092567|emb|CAC29731.1|
putative GTP cyclohydrolase I [Mycobacterium leprae]
Length = 205
Score = 100 bits (250), Expect = 2e-20
Identities = 56/113 (49%), Positives = 69/113 (60%), Gaps = 1/113 (0%)
Frame = -3
Query: 630 LPFWSQCEHHLLPFHGVVHIGYFISDGFKPITKSLLQSMVHFYGFKLQVQERLTRQIAET 451
+P +S CEHHL+ FHGV HIGY + S + +V Y + QVQERLT QIA+
Sbjct: 87 IPMYSTCEHHLVSFHGVAHIGYLPGADGRVTGLSKIARLVDLYAKRPQVQERLTSQIADA 146
Query: 450 -VSPLLGGDVIVVVEASHTCMISRGIEKFGSSTATIAVLGRFSTDIAARASFL 295
VS L VI+VVEA H CM RG+ K G+ T T AV G+F TD A+RA L
Sbjct: 147 LVSKLDPRGVIIVVEAEHLCMAMRGVRKPGAITTTSAVRGQFKTDAASRAEAL 199
>emb|CAC86189.1| GTP cyclohydrolase I [Danio rerio]
Length = 240
Score = 99.8 bits (247), Expect = 4e-20
Identities = 57/121 (47%), Positives = 74/121 (61%), Gaps = 1/121 (0%)
Frame = -3
Query: 654 EKIYSEVNLPFWSQCEHHLLPFHGVVHIGYFISDGFKPITKSLLQSMVHFYGFKLQVQER 475
E++ ++ +S CEHHL+PF G VHIGY S K + S L +V Y +LQVQER
Sbjct: 115 EELVIVKDIDMFSLCEHHLVPFFGKVHIGYLPSK--KVVGLSKLARIVEIYSRRLQVQER 172
Query: 474 LTRQIAETVSPLLGG-DVIVVVEASHTCMISRGIEKFGSSTATIAVLGRFSTDIAARASF 298
LT+QIA +S L V VV+EA+H CM+ RG++K S T T A+LG F D AR F
Sbjct: 173 LTKQIAMAISEALQPVGVAVVIEAAHMCMVMRGVQKMNSHTVTSAMLGVFREDPKAREEF 232
Query: 297 L 295
L
Sbjct: 233 L 233
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 560,726,489
Number of Sequences: 1393205
Number of extensions: 11334245
Number of successful extensions: 27122
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 25873
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26802
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34062062287
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)