Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000839A_C01 KMC000839A_c01
(575 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
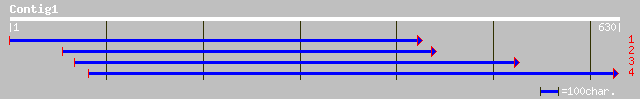
ref|NP_199623.1| unknown protein; protein id: At5g48120.1 [Arabi... 62 7e-09
sp|P77809|G6PD_ACTAC Glucose-6-phosphate 1-dehydrogenase (G6PD) ... 33 2.6
gb|AAO51863.1| hypothetical protein [Dictyostelium discoideum] 33 3.4
ref|ZP_00118198.1| hypothetical protein [Cytophaga hutchinsonii] 32 4.4
gb|ZP_00023849.1| hypothetical protein [Ralstonia metallidurans] 32 4.4
>ref|NP_199623.1| unknown protein; protein id: At5g48120.1 [Arabidopsis thaliana]
Length = 1152
Score = 61.6 bits (148), Expect = 7e-09
Identities = 38/110 (34%), Positives = 58/110 (52%), Gaps = 22/110 (20%)
Frame = -1
Query: 362 RSFLYRAFAHIISDTPLIVIVSEAKKLIPVLLNCLSMLTEDIQDKDILYGLLLVLSGILT 183
R+ L+ A AH+IS+ P+ VI+ KKL P++L LS+L+ D +K+ L+ LLLVLSG LT
Sbjct: 1001 RTMLHVALAHVISNVPVTVILDNTKKLQPLILEGLSVLSLDSVEKETLFSLLLVLSGTLT 1060
Query: 182 EK----------------------NGQEAVTENAYIIINCLIKFVDYPRK 99
+ NG + V E + I CL+ ++ P +
Sbjct: 1061 DTKASSFFPTELILESSNDNMSTLNGMQVVRETS---IQCLVALLELPHR 1107
>sp|P77809|G6PD_ACTAC Glucose-6-phosphate 1-dehydrogenase (G6PD)
gi|1651208|dbj|BAA13554.1| glucose-6-phosphate
dehydrogenase [Actinobacillus actinomycetemcomitans]
Length = 494
Score = 33.1 bits (74), Expect = 2.6
Identities = 20/94 (21%), Positives = 44/94 (46%), Gaps = 7/94 (7%)
Frame = -1
Query: 380 ALMDWKRSFLYRAFAHIISDTPLIVIVSEAKKLIPVLLNCLSMLTEDIQDKDILYGLLLV 201
A+ D ++ L + A + + P+I+ + + + +L+CL LT++ + +++ G +
Sbjct: 234 AMRDMFQNHLLQVLAMVAMEPPVIINANSMRDEVAKVLHCLRPLTQEDVEHNLVLGQYVA 293
Query: 200 -------LSGILTEKNGQEAVTENAYIIINCLIK 120
+ G L EK T Y+ + C I+
Sbjct: 294 GEVDGEWVKGYLEEKGVPPYSTTETYMALRCEIE 327
>gb|AAO51863.1| hypothetical protein [Dictyostelium discoideum]
Length = 858
Score = 32.7 bits (73), Expect = 3.4
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Frame = +1
Query: 427 MPRHYSVRTISLPLSAPLHPCNTSVL-QTHIFVLNITF 537
MPRH V S AP+H NTSV+ Q +F L+I F
Sbjct: 70 MPRHLIVSRFSSNYFAPIHLINTSVVFQAFLFALHIVF 107
>ref|ZP_00118198.1| hypothetical protein [Cytophaga hutchinsonii]
Length = 154
Score = 32.3 bits (72), Expect = 4.4
Identities = 23/63 (36%), Positives = 29/63 (45%), Gaps = 6/63 (9%)
Frame = -1
Query: 350 YRAFAHIISDTPLIVIVSEAKKLIPVLLNCLSMLTEDIQDKDI------LYGLLLVLSGI 189
+R F H I DTP I + AK + +L L L E I DI L L VL+G
Sbjct: 14 HRTFKHPILDTPAIPAIERAKLRVALLAEELKELEEAIAQNDIVEAADALCDLQYVLAGA 73
Query: 188 LTE 180
+ E
Sbjct: 74 VLE 76
>gb|ZP_00023849.1| hypothetical protein [Ralstonia metallidurans]
Length = 325
Score = 32.3 bits (72), Expect = 4.4
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 1/65 (1%)
Frame = -1
Query: 296 EAKKLIPVLLNCLSMLTEDIQDKDILYGLLL-VLSGILTEKNGQEAVTENAYIIINCLIK 120
EA++ + LL ++ L + ++K + GLLL I E NG ENA++ ++ +
Sbjct: 168 EAEQRVARLLTTIARLQQKAREKGAIGGLLLGATDSITAEPNGDVGSIENAWLAVSLALA 227
Query: 119 FVDYP 105
YP
Sbjct: 228 KQAYP 232
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 475,504,183
Number of Sequences: 1393205
Number of extensions: 9674076
Number of successful extensions: 22506
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 21960
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22502
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)