Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000834A_C01 KMC000834A_c01
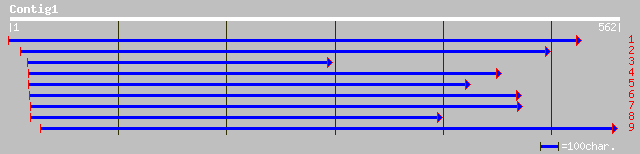
(562 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB16489.1| P0665D10.14 [Oryza sativa (japonica cultivar-gro... 94 9e-19
ref|NP_196078.1| putative protein; protein id: At5g04580.1 [Arab... 88 8e-17
gb|AAM77215.1| DEMETER protein [Arabidopsis thaliana] 88 8e-17
dbj|BAC42629.1| unknown protein [Arabidopsis thaliana] gi|289509... 88 8e-17
gb|AAL69364.1|AF462201_1 unknown [Narcissus pseudonarcissus] 82 6e-15
>dbj|BAB16489.1| P0665D10.14 [Oryza sativa (japonica cultivar-group)]
gi|21327943|dbj|BAC00536.1| P0489G09.3 [Oryza sativa
(japonica cultivar-group)]
Length = 274
Score = 94.4 bits (233), Expect = 9e-19
Identities = 41/55 (74%), Positives = 47/55 (84%)
Frame = -1
Query: 562 GTSVPSIFKGLTTPEIQQCFWRGFVCVRAFERGTRAPRPLKGRLHFPVSKMPKNK 398
GTS+P+IFKGLTT EIQ CFWRGFVCVR F+R +RAPRPL RLHFP SK+ +NK
Sbjct: 208 GTSIPTIFKGLTTEEIQHCFWRGFVCVRGFDRTSRAPRPLYARLHFPASKITRNK 262
>ref|NP_196078.1| putative protein; protein id: At5g04580.1 [Arabidopsis thaliana]
gi|11290070|pir||T48454 hypothetical protein T32M21.180
- Arabidopsis thaliana gi|7406462|emb|CAB85564.1|
putative protein [Arabidopsis thaliana]
Length = 234
Score = 87.8 bits (216), Expect = 8e-17
Identities = 39/55 (70%), Positives = 45/55 (80%)
Frame = -1
Query: 562 GTSVPSIFKGLTTPEIQQCFWRGFVCVRAFERGTRAPRPLKGRLHFPVSKMPKNK 398
GTSV SIF+GL+T +IQ CFW+GFVCVR FE+ TRAPRPL RLHFP SK+ NK
Sbjct: 179 GTSVTSIFRGLSTEQIQFCFWKGFVCVRGFEQKTRAPRPLMARLHFPASKLKNNK 233
>gb|AAM77215.1| DEMETER protein [Arabidopsis thaliana]
Length = 1729
Score = 87.8 bits (216), Expect = 8e-17
Identities = 39/55 (70%), Positives = 45/55 (80%)
Frame = -1
Query: 562 GTSVPSIFKGLTTPEIQQCFWRGFVCVRAFERGTRAPRPLKGRLHFPVSKMPKNK 398
GTSV SIF+GL+T +IQ CFW+GFVCVR FE+ TRAPRPL RLHFP SK+ NK
Sbjct: 1674 GTSVTSIFRGLSTEQIQFCFWKGFVCVRGFEQKTRAPRPLMARLHFPASKLKNNK 1728
>dbj|BAC42629.1| unknown protein [Arabidopsis thaliana] gi|28950995|gb|AAO63421.1|
At5g04570 [Arabidopsis thaliana]
Length = 416
Score = 87.8 bits (216), Expect = 8e-17
Identities = 39/55 (70%), Positives = 45/55 (80%)
Frame = -1
Query: 562 GTSVPSIFKGLTTPEIQQCFWRGFVCVRAFERGTRAPRPLKGRLHFPVSKMPKNK 398
GTSV SIF+GL+T +IQ CFW+GFVCVR FE+ TRAPRPL RLHFP SK+ NK
Sbjct: 361 GTSVTSIFRGLSTEQIQFCFWKGFVCVRGFEQKTRAPRPLMARLHFPASKLKNNK 415
>gb|AAL69364.1|AF462201_1 unknown [Narcissus pseudonarcissus]
Length = 53
Score = 81.6 bits (200), Expect = 6e-15
Identities = 37/51 (72%), Positives = 40/51 (77%)
Frame = -1
Query: 550 PSIFKGLTTPEIQQCFWRGFVCVRAFERGTRAPRPLKGRLHFPVSKMPKNK 398
PSIFKGLTT IQ CFWRGFVCVR F+R RAP+PL RLHFP SK +NK
Sbjct: 1 PSIFKGLTTEGIQHCFWRGFVCVRGFDRMMRAPKPLFARLHFPASKGSRNK 51
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 506,090,308
Number of Sequences: 1393205
Number of extensions: 11162981
Number of successful extensions: 30753
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 29287
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30678
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)