Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000831A_C01 KMC000831A_c01
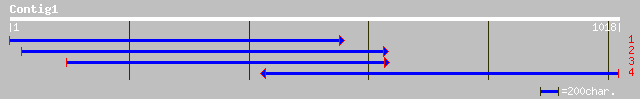
(998 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_179734.1| coatomer alpha subunit; protein id: At2g21390.1... 518 e-146
gb|AAK18834.1|AC082645_4 putative alpha-coat protein [Oryza sativa] 515 e-145
gb|AAK18837.1|AC082645_7 putative alpha-coat protein [Oryza sativa] 513 e-144
ref|NP_176393.1| coatomer alpha subunit, putative; protein id: A... 513 e-144
ref|NP_004362.1| coatomer protein complex, subunit alpha; alpha ... 449 e-125
>ref|NP_179734.1| coatomer alpha subunit; protein id: At2g21390.1 [Arabidopsis
thaliana] gi|25301669|pir||F84600 coatomer alpha subunit
[imported] - Arabidopsis thaliana
gi|4567286|gb|AAD23699.1| coatomer alpha subunit
[Arabidopsis thaliana]
Length = 1218
Score = 518 bits (1334), Expect = e-146
Identities = 239/258 (92%), Positives = 252/258 (97%)
Frame = +3
Query: 222 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFH 401
MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLID+FDEH+GPVRGVHFH
Sbjct: 1 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDRFDEHEGPVRGVHFH 60
Query: 402 HSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIW 581
+SQPLFVSGGDDYKIKVWNYK HRCLFTLLGHLDYIRTVQFHHE+PWIVSASDDQTIRIW
Sbjct: 61 NSQPLFVSGGDDYKIKVWNYKTHRCLFTLLGHLDYIRTVQFHHENPWIVSASDDQTIRIW 120
Query: 582 NWQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDIGSLKRKNASPADDILR 761
NWQSRTCISVLTGHNHYVMCA FHP+EDLVVSASLDQTVRVWDIG+LK+K+ASPADD++R
Sbjct: 121 NWQSRTCISVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWDIGALKKKSASPADDLMR 180
Query: 762 LSQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPALPLIVSAADDRQVKLWRMNDTKAWE 941
SQMN+DLFGGVDA+VKYVLEGHDRGVNWASFHP LPLIVS ADDRQVKLWRMN+TKAWE
Sbjct: 181 FSQMNSDLFGGVDAIVKYVLEGHDRGVNWASFHPTLPLIVSGADDRQVKLWRMNETKAWE 240
Query: 942 VDTLRGHMNNVSCVMFHA 995
VDTLRGHMNNVS VMFHA
Sbjct: 241 VDTLRGHMNNVSSVMFHA 258
Score = 84.7 bits (208), Expect = 2e-15
Identities = 48/181 (26%), Positives = 84/181 (45%), Gaps = 29/181 (16%)
Frame = +3
Query: 255 VKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGD 434
++ + FH + PWI+++ I++W+++ T I H+ V FH + L VS
Sbjct: 96 IRTVQFHHENPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCASFHPKEDLVVSASL 155
Query: 435 DYKIKVWNY------------------KLHRCLF---------TLLGHLDYIRTVQFHHE 533
D ++VW+ +++ LF L GH + FH
Sbjct: 156 DQTVRVWDIGALKKKSASPADDLMRFSQMNSDLFGGVDAIVKYVLEGHDRGVNWASFHPT 215
Query: 534 SPWIVSASDDQTIRIWNWQSRTC--ISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVW 707
P IVS +DD+ +++W + L GH + V +FH ++D++VS S D+++RVW
Sbjct: 216 LPLIVSGADDRQVKLWRMNETKAWEVDTLRGHMNNVSSVMFHAKQDIIVSNSEDKSIRVW 275
Query: 708 D 710
D
Sbjct: 276 D 276
Score = 70.9 bits (172), Expect = 3e-11
Identities = 48/187 (25%), Positives = 82/187 (43%), Gaps = 31/187 (16%)
Frame = +3
Query: 210 QSKKMLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDK---------- 359
QS+ ++ ++ V SFHPK ++++ +++WD +G L K
Sbjct: 123 QSRTCISVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWD--IGALKKKSASPADDLMR 180
Query: 360 -------------------FDEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRC-- 476
+ HD V FH + PL VSG DD ++K+W +
Sbjct: 181 FSQMNSDLFGGVDAIVKYVLEGHDRGVNWASFHPTLPLIVSGADDRQVKLWRMNETKAWE 240
Query: 477 LFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHP 656
+ TL GH++ + +V FH + IVS S+D++IR+W+ RT I + HP
Sbjct: 241 VDTLRGHMNNVSSVMFHAKQDIIVSNSEDKSIRVWDATKRTGIQTFRREHDRFWILAVHP 300
Query: 657 REDLVVS 677
+L+ +
Sbjct: 301 EINLLAA 307
Score = 41.2 bits (95), Expect = 0.025
Identities = 30/110 (27%), Positives = 44/110 (39%), Gaps = 4/110 (3%)
Frame = +3
Query: 255 VKGLSFHPKRPWILASLHSGVIQLWDYRMGTL----IDKFDEHDGPVRGVHFHHSQPLFV 422
V SFHP P I++ ++LW RM +D H V V FH Q + V
Sbjct: 207 VNWASFHPTLPLIVSGADDRQVKLW--RMNETKAWEVDTLRGHMNNVSSVMFHAKQDIIV 264
Query: 423 SGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTI 572
S +D I+VW+ + T D + H E + + D+ I
Sbjct: 265 SNSEDKSIRVWDATKRTGIQTFRREHDRFWILAVHPEINLLAAGHDNGMI 314
>gb|AAK18834.1|AC082645_4 putative alpha-coat protein [Oryza sativa]
Length = 1218
Score = 515 bits (1327), Expect = e-145
Identities = 237/258 (91%), Positives = 249/258 (95%)
Frame = +3
Query: 222 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFH 401
MLTKFETKSNRVKGLSFHP+RPWILASLHSGVIQ+WDYRMGTL+D+FDEHDGPVRGVHFH
Sbjct: 1 MLTKFETKSNRVKGLSFHPRRPWILASLHSGVIQMWDYRMGTLLDRFDEHDGPVRGVHFH 60
Query: 402 HSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIW 581
+QPLFVSGGDDYKIKVWNYK HRCLFTL GHLDYIRTVQFHHE PWIVSASDDQTIRIW
Sbjct: 61 ATQPLFVSGGDDYKIKVWNYKTHRCLFTLHGHLDYIRTVQFHHEYPWIVSASDDQTIRIW 120
Query: 582 NWQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDIGSLKRKNASPADDILR 761
NWQSRTC++VLTGHNHYVMCA FHP+EDLVVSASLDQTVRVWDIG+L++K SPADDILR
Sbjct: 121 NWQSRTCVAVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWDIGALRKKTVSPADDILR 180
Query: 762 LSQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPALPLIVSAADDRQVKLWRMNDTKAWE 941
L+QMNTDLFGGVDAVVKYVLEGHDRGVNWASFHP LPLIVS ADDRQVKLWRMNDTKAWE
Sbjct: 181 LTQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPTLPLIVSGADDRQVKLWRMNDTKAWE 240
Query: 942 VDTLRGHMNNVSCVMFHA 995
VDTLRGHMNNVSCVMFHA
Sbjct: 241 VDTLRGHMNNVSCVMFHA 258
Score = 86.3 bits (212), Expect = 7e-16
Identities = 46/181 (25%), Positives = 82/181 (44%), Gaps = 29/181 (16%)
Frame = +3
Query: 255 VKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGD 434
++ + FH + PWI+++ I++W+++ T + H+ V FH + L VS
Sbjct: 96 IRTVQFHHEYPWIVSASDDQTIRIWNWQSRTCVAVLTGHNHYVMCASFHPKEDLVVSASL 155
Query: 435 DYKIKVWNYKLHR---------------------------CLFTLLGHLDYIRTVQFHHE 533
D ++VW+ R + L GH + FH
Sbjct: 156 DQTVRVWDIGALRKKTVSPADDILRLTQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPT 215
Query: 534 SPWIVSASDDQTIRIWNWQSRTC--ISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVW 707
P IVS +DD+ +++W + L GH + V C +FH ++D++VS S D+++R+W
Sbjct: 216 LPLIVSGADDRQVKLWRMNDTKAWEVDTLRGHMNNVSCVMFHAKQDIIVSNSEDKSIRIW 275
Query: 708 D 710
D
Sbjct: 276 D 276
Score = 68.9 bits (167), Expect = 1e-10
Identities = 49/187 (26%), Positives = 80/187 (42%), Gaps = 31/187 (16%)
Frame = +3
Query: 210 QSKKMLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDK---------- 359
QS+ + ++ V SFHPK ++++ +++WD +G L K
Sbjct: 123 QSRTCVAVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWD--IGALRKKTVSPADDILR 180
Query: 360 -------------------FDEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRC-- 476
+ HD V FH + PL VSG DD ++K+W +
Sbjct: 181 LTQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPTLPLIVSGADDRQVKLWRMNDTKAWE 240
Query: 477 LFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHP 656
+ TL GH++ + V FH + IVS S+D++IRIW+ RT I + HP
Sbjct: 241 VDTLRGHMNNVSCVMFHAKQDIIVSNSEDKSIRIWDATKRTGIQTFRREHDRFWILSAHP 300
Query: 657 REDLVVS 677
+L+ +
Sbjct: 301 EMNLLAA 307
>gb|AAK18837.1|AC082645_7 putative alpha-coat protein [Oryza sativa]
Length = 1218
Score = 513 bits (1321), Expect = e-144
Identities = 235/258 (91%), Positives = 249/258 (96%)
Frame = +3
Query: 222 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFH 401
MLTKFETKSNRVKGLSFHP+RPWILASLHSGVIQ+WDYRMGTL+D+FDEHDGPVRGVHFH
Sbjct: 1 MLTKFETKSNRVKGLSFHPRRPWILASLHSGVIQMWDYRMGTLLDRFDEHDGPVRGVHFH 60
Query: 402 HSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIW 581
+QPLFVSGGDDYKIKVWNYK HRCLFTL GHLDYIRTVQFHHE PWIVSASDDQTIRIW
Sbjct: 61 ATQPLFVSGGDDYKIKVWNYKTHRCLFTLHGHLDYIRTVQFHHECPWIVSASDDQTIRIW 120
Query: 582 NWQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDIGSLKRKNASPADDILR 761
NWQSRTC++VLTGHNHYVMCA FHP+EDLVVSASLDQTVRVWDI +L++K+ SPADDILR
Sbjct: 121 NWQSRTCVAVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWDISALRKKSVSPADDILR 180
Query: 762 LSQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPALPLIVSAADDRQVKLWRMNDTKAWE 941
L+QMNTDLFGGVDAVVKYVLEGHDRGVNWASFHP LPLIVS ADDRQVK+WRMNDTKAWE
Sbjct: 181 LTQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPTLPLIVSGADDRQVKIWRMNDTKAWE 240
Query: 942 VDTLRGHMNNVSCVMFHA 995
VDTLRGHMNNVSCVMFHA
Sbjct: 241 VDTLRGHMNNVSCVMFHA 258
Score = 87.4 bits (215), Expect = 3e-16
Identities = 47/181 (25%), Positives = 82/181 (44%), Gaps = 29/181 (16%)
Frame = +3
Query: 255 VKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGD 434
++ + FH + PWI+++ I++W+++ T + H+ V FH + L VS
Sbjct: 96 IRTVQFHHECPWIVSASDDQTIRIWNWQSRTCVAVLTGHNHYVMCASFHPKEDLVVSASL 155
Query: 435 DYKIKVWNYKLHR---------------------------CLFTLLGHLDYIRTVQFHHE 533
D ++VW+ R + L GH + FH
Sbjct: 156 DQTVRVWDISALRKKSVSPADDILRLTQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPT 215
Query: 534 SPWIVSASDDQTIRIWNWQSRTC--ISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVW 707
P IVS +DD+ ++IW + L GH + V C +FH ++D++VS S D+++R+W
Sbjct: 216 LPLIVSGADDRQVKIWRMNDTKAWEVDTLRGHMNNVSCVMFHAKQDIIVSNSEDKSIRIW 275
Query: 708 D 710
D
Sbjct: 276 D 276
Score = 68.9 bits (167), Expect = 1e-10
Identities = 49/185 (26%), Positives = 82/185 (43%), Gaps = 29/185 (15%)
Frame = +3
Query: 210 QSKKMLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDY------------------ 335
QS+ + ++ V SFHPK ++++ +++WD
Sbjct: 123 QSRTCVAVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWDISALRKKSVSPADDILRLT 182
Query: 336 RMGT-------LIDKF--DEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRC--LF 482
+M T + K+ + HD V FH + PL VSG DD ++K+W + +
Sbjct: 183 QMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPTLPLIVSGADDRQVKIWRMNDTKAWEVD 242
Query: 483 TLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHPRE 662
TL GH++ + V FH + IVS S+D++IRIW+ RT I + HP
Sbjct: 243 TLRGHMNNVSCVMFHAKQDIIVSNSEDKSIRIWDATKRTGIQTFRREHDRFWILSAHPEM 302
Query: 663 DLVVS 677
+L+ +
Sbjct: 303 NLLAA 307
>ref|NP_176393.1| coatomer alpha subunit, putative; protein id: At1g62020.1
[Arabidopsis thaliana] gi|7444289|pir||T02146 coatomer
complex alpha chain homolog F8K4.21 - Arabidopsis
thaliana gi|3367534|gb|AAC28519.1| Strong similarity to
coatamer alpha subunit (HEPCOP) homolog gb|U24105 from
Homo sapiens. [Arabidopsis thaliana]
Length = 1216
Score = 513 bits (1320), Expect = e-144
Identities = 236/258 (91%), Positives = 250/258 (96%)
Frame = +3
Query: 222 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFH 401
MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLID+FDEH+GPVRGVHFH
Sbjct: 1 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDRFDEHEGPVRGVHFH 60
Query: 402 HSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIW 581
+SQPLFVSGGDDYKIKVWNYK HRCLFTLLGHLDYIRTVQFHHE PWIVSASDDQTIRIW
Sbjct: 61 NSQPLFVSGGDDYKIKVWNYKNHRCLFTLLGHLDYIRTVQFHHEYPWIVSASDDQTIRIW 120
Query: 582 NWQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDIGSLKRKNASPADDILR 761
NWQSRTC+SVLTGHNHYVMCA FHP+EDLVVSASLDQTVRVWDIG+L++K SPADDI+R
Sbjct: 121 NWQSRTCVSVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWDIGALRKKTVSPADDIMR 180
Query: 762 LSQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPALPLIVSAADDRQVKLWRMNDTKAWE 941
L+QMN+DLFGGVDA+VKYVLEGHDRGVNWA+FHP LPLIVS ADDRQVKLWRMN+TKAWE
Sbjct: 181 LTQMNSDLFGGVDAIVKYVLEGHDRGVNWAAFHPTLPLIVSGADDRQVKLWRMNETKAWE 240
Query: 942 VDTLRGHMNNVSCVMFHA 995
VDTLRGHMNNVS VMFHA
Sbjct: 241 VDTLRGHMNNVSSVMFHA 258
Score = 83.2 bits (204), Expect = 6e-15
Identities = 46/181 (25%), Positives = 81/181 (44%), Gaps = 29/181 (16%)
Frame = +3
Query: 255 VKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGD 434
++ + FH + PWI+++ I++W+++ T + H+ V FH + L VS
Sbjct: 96 IRTVQFHHEYPWIVSASDDQTIRIWNWQSRTCVSVLTGHNHYVMCASFHPKEDLVVSASL 155
Query: 435 DYKIKVWNYKLHR---------------------------CLFTLLGHLDYIRTVQFHHE 533
D ++VW+ R + L GH + FH
Sbjct: 156 DQTVRVWDIGALRKKTVSPADDIMRLTQMNSDLFGGVDAIVKYVLEGHDRGVNWAAFHPT 215
Query: 534 SPWIVSASDDQTIRIWNWQSRTC--ISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVW 707
P IVS +DD+ +++W + L GH + V +FH ++D++VS S D+++RVW
Sbjct: 216 LPLIVSGADDRQVKLWRMNETKAWEVDTLRGHMNNVSSVMFHAKQDIIVSNSEDKSIRVW 275
Query: 708 D 710
D
Sbjct: 276 D 276
Score = 69.7 bits (169), Expect = 7e-11
Identities = 47/187 (25%), Positives = 82/187 (43%), Gaps = 31/187 (16%)
Frame = +3
Query: 210 QSKKMLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDK---------- 359
QS+ ++ ++ V SFHPK ++++ +++WD +G L K
Sbjct: 123 QSRTCVSVLTGHNHYVMCASFHPKEDLVVSASLDQTVRVWD--IGALRKKTVSPADDIMR 180
Query: 360 -------------------FDEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRC-- 476
+ HD V FH + PL VSG DD ++K+W +
Sbjct: 181 LTQMNSDLFGGVDAIVKYVLEGHDRGVNWAAFHPTLPLIVSGADDRQVKLWRMNETKAWE 240
Query: 477 LFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHP 656
+ TL GH++ + +V FH + IVS S+D++IR+W+ RT + + HP
Sbjct: 241 VDTLRGHMNNVSSVMFHAKQDIIVSNSEDKSIRVWDATKRTGLQTFRREHDRFWILAVHP 300
Query: 657 REDLVVS 677
+L+ +
Sbjct: 301 EMNLLAA 307
Score = 40.8 bits (94), Expect = 0.033
Identities = 30/110 (27%), Positives = 43/110 (38%), Gaps = 4/110 (3%)
Frame = +3
Query: 255 VKGLSFHPKRPWILASLHSGVIQLWDYRMGTL----IDKFDEHDGPVRGVHFHHSQPLFV 422
V +FHP P I++ ++LW RM +D H V V FH Q + V
Sbjct: 207 VNWAAFHPTLPLIVSGADDRQVKLW--RMNETKAWEVDTLRGHMNNVSSVMFHAKQDIIV 264
Query: 423 SGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTI 572
S +D I+VW+ L T D + H E + + D I
Sbjct: 265 SNSEDKSIRVWDATKRTGLQTFRREHDRFWILAVHPEMNLLAAGHDSGMI 314
>ref|NP_004362.1| coatomer protein complex, subunit alpha; alpha coat protein; xenin
[Homo sapiens] gi|1705996|sp|P53621|COPA_HUMAN Coatomer
alpha subunit (Alpha-coat protein) (Alpha-COP) (HEPCOP)
(HEP-COP) [Contains: Xenin (Xenopsin-related peptide);
Proxenin] gi|2144979|pir||ERHUAH coatomer complex alpha
chain homolog - human gi|1002369|gb|AAB70879.1| coatomer
protein
Length = 1224
Score = 449 bits (1155), Expect = e-125
Identities = 209/260 (80%), Positives = 226/260 (86%), Gaps = 3/260 (1%)
Frame = +3
Query: 222 MLTKFETKSNRVKGLSFHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFH 401
MLTKFETKS RVKGLSFHPKRPWIL SLH+GVIQLWDYRM TLIDKFDEHDGPVRG+ FH
Sbjct: 1 MLTKFETKSARVKGLSFHPKRPWILTSLHNGVIQLWDYRMCTLIDKFDEHDGPVRGIDFH 60
Query: 402 HSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIW 581
QPLFVSGGDDYKIKVWNYKL RCLFTLLGHLDYIRT FHHE PWI+SASDDQTIR+W
Sbjct: 61 KQQPLFVSGGDDYKIKVWNYKLRRCLFTLLGHLDYIRTTFFHHEYPWILSASDDQTIRVW 120
Query: 582 NWQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVWDIGSLKRKNASPA---DD 752
NWQSRTC+ VLTGHNHYVMCA FHP EDLVVSASLDQTVRVWDI L++KN SP D
Sbjct: 121 NWQSRTCVCVLTGHNHYVMCAQFHPTEDLVVSASLDQTVRVWDISGLRKKNLSPGAVESD 180
Query: 753 ILRLSQMNTDLFGGVDAVVKYVLEGHDRGVNWASFHPALPLIVSAADDRQVKLWRMNDTK 932
+ ++ DLFG DAVVK+VLEGHDRGVNWA+FHP +PLIVS ADDRQVK+WRMN++K
Sbjct: 181 VRGIT--GVDLFGTTDAVVKHVLEGHDRGVNWAAFHPTMPLIVSGADDRQVKIWRMNESK 238
Query: 933 AWEVDTLRGHMNNVSCVMFH 992
AWEVDT RGH NNVSC +FH
Sbjct: 239 AWEVDTCRGHYNNVSCAVFH 258
Score = 88.6 bits (218), Expect = 1e-16
Identities = 55/182 (30%), Positives = 86/182 (47%), Gaps = 34/182 (18%)
Frame = +3
Query: 270 FHPKRPWILASLHSGVIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGDDYKIK 449
FH + PWIL++ I++W+++ T + H+ V FH ++ L VS D ++
Sbjct: 101 FHHEYPWILSASDDQTIRVWNWQSRTCVCVLTGHNHYVMCAQFHPTEDLVVSASLDQTVR 160
Query: 450 VWNYKLHRC-------------------LF---------TLLGHLDYIRTVQFHHESPWI 545
VW+ R LF L GH + FH P I
Sbjct: 161 VWDISGLRKKNLSPGAVESDVRGITGVDLFGTTDAVVKHVLEGHDRGVNWAAFHPTMPLI 220
Query: 546 VSASDDQTIRIWN------WQSRTCISVLTGHNHYVMCALFHPREDLVVSASLDQTVRVW 707
VS +DD+ ++IW W+ TC GH + V CA+FHPR++L++S S D+++RVW
Sbjct: 221 VSGADDRQVKIWRMNESKAWEVDTC----RGHYNNVSCAVFHPRQELILSNSEDKSIRVW 276
Query: 708 DI 713
D+
Sbjct: 277 DM 278
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 908,959,957
Number of Sequences: 1393205
Number of extensions: 22278258
Number of successful extensions: 287630
Number of sequences better than 10.0: 3292
Number of HSP's better than 10.0 without gapping: 101157
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 194097
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 57393820016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)