Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
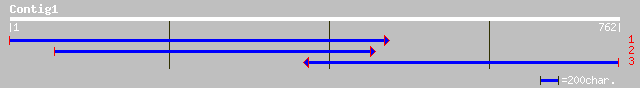
Query= KMC000822A_C01 KMC000822A_c01
(762 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_179370.1| putative vacuolar sorting protein 35; protein i... 339 2e-92
pir||T08858 vacuolar protein-sorting protein homolog A_TM017A05.... 339 2e-92
gb|AAO72382.1| putative vacuolar protein sorting-associated prot... 335 4e-91
gb|AAO37536.1| putative vacuolar sorting-associated protein, 3'-... 328 2e-89
ref|NP_177713.1| vacuolor sorting protein 35, putative; protein ... 300 1e-80
>ref|NP_179370.1| putative vacuolar sorting protein 35; protein id: At2g17790.1
[Arabidopsis thaliana] gi|25336361|pir||E84556 probable
vacuolar sorting-associated protein [imported] -
Arabidopsis thaliana
Length = 830
Score = 339 bits (870), Expect = 2e-92
Identities = 167/206 (81%), Positives = 187/206 (90%), Gaps = 1/206 (0%)
Frame = +3
Query: 147 MMLDGTEDEEKFLAAGIAGLQQNSFYMHRALDSNNLRDALKYSAQMLSELRTSKLSPHKY 326
M+ DG+EDEEK+LAAG A +QN+FYM RA+DSNNL+DALKYSAQMLSELRTSKLSPHKY
Sbjct: 1 MIADGSEDEEKWLAAGAAAFKQNAFYMQRAIDSNNLKDALKYSAQMLSELRTSKLSPHKY 60
Query: 327 YELYMRAFDQLRKLEMFFEEEARRGCSIIDLYELVQHAGNILPRLYLLCTVGSVYIKSKE 506
Y+LYMRAFD+LRKLE+FF EE RRGCS+I+LYELVQHAGNILPRLYLLCT GSVYIK+KE
Sbjct: 61 YDLYMRAFDELRKLEIFFMEETRRGCSVIELYELVQHAGNILPRLYLLCTAGSVYIKTKE 120
Query: 507 APAKDVLKDLVEMCRGIQHPVRGLFLRSYLSQVSRDKLPDIGSEYEGDADTVADAVEFVL 686
APAK++LKDLVEMCRGIQHP+RGLFLRSYL+Q+SRDKLPDIGSEYEGDADTV DAVEFVL
Sbjct: 121 APAKEILKDLVEMCRGIQHPLRGLFLRSYLAQISRDKLPDIGSEYEGDADTVIDAVEFVL 180
Query: 687 QNFTEMNNFGCGCNIR-PAREKEKRE 761
NFTEMN + PAREKE+RE
Sbjct: 181 LNFTEMNKLWVRMQHQGPAREKERRE 206
>pir||T08858 vacuolar protein-sorting protein homolog A_TM017A05.7 - Arabidopsis
thaliana
Length = 848
Score = 339 bits (870), Expect = 2e-92
Identities = 167/206 (81%), Positives = 187/206 (90%), Gaps = 1/206 (0%)
Frame = +3
Query: 147 MMLDGTEDEEKFLAAGIAGLQQNSFYMHRALDSNNLRDALKYSAQMLSELRTSKLSPHKY 326
M+ DG+EDEEK+LAAG A +QN+FYM RA+DSNNL+DALKYSAQMLSELRTSKLSPHKY
Sbjct: 1 MIADGSEDEEKWLAAGAAAFKQNAFYMQRAIDSNNLKDALKYSAQMLSELRTSKLSPHKY 60
Query: 327 YELYMRAFDQLRKLEMFFEEEARRGCSIIDLYELVQHAGNILPRLYLLCTVGSVYIKSKE 506
Y+LYMRAFD+LRKLE+FF EE RRGCS+I+LYELVQHAGNILPRLYLLCT GSVYIK+KE
Sbjct: 61 YDLYMRAFDELRKLEIFFMEETRRGCSVIELYELVQHAGNILPRLYLLCTAGSVYIKTKE 120
Query: 507 APAKDVLKDLVEMCRGIQHPVRGLFLRSYLSQVSRDKLPDIGSEYEGDADTVADAVEFVL 686
APAK++LKDLVEMCRGIQHP+RGLFLRSYL+Q+SRDKLPDIGSEYEGDADTV DAVEFVL
Sbjct: 121 APAKEILKDLVEMCRGIQHPLRGLFLRSYLAQISRDKLPDIGSEYEGDADTVIDAVEFVL 180
Query: 687 QNFTEMNNFGCGCNIR-PAREKEKRE 761
NFTEMN + PAREKE+RE
Sbjct: 181 LNFTEMNKLWVRMQHQGPAREKERRE 206
>gb|AAO72382.1| putative vacuolar protein sorting-associated protein [Oryza sativa
(japonica cultivar-group)]
Length = 793
Score = 335 bits (859), Expect = 4e-91
Identities = 166/202 (82%), Positives = 184/202 (90%), Gaps = 2/202 (0%)
Frame = +3
Query: 159 GTEDEEKFLAAGIAGLQQNSFYMHRALDSNNLRDALKYSAQMLSELRTSKLSPHKYYELY 338
G +DEE++LA GIAG+QQN+FYMHRALDSNNL+DALKYSAQMLSELRTS+LSPHKYY+LY
Sbjct: 6 GADDEERWLAEGIAGVQQNAFYMHRALDSNNLKDALKYSAQMLSELRTSRLSPHKYYDLY 65
Query: 339 MRAFDQLRKLEMFFEEEARRG-CSIIDLYELVQHAGNILPRLYLLCTVGSVYIKSKEAPA 515
MRAFD++RKLEMFF EE RRG CS++DLYELVQHAGN+LPRLYLLCTVGSVYIKSKEAPA
Sbjct: 66 MRAFDEMRKLEMFFREETRRGSCSVVDLYELVQHAGNVLPRLYLLCTVGSVYIKSKEAPA 125
Query: 516 KDVLKDLVEMCRGIQHPVRGLFLRSYLSQVSRDKLPDIGSEYEGDADTVADAVEFVLQNF 695
KDVLKDLVEMCRGIQHP+RGLFLRSYLSQ+SRDKLPDIGSEYEGDAD++ AVEFVLQNF
Sbjct: 126 KDVLKDLVEMCRGIQHPLRGLFLRSYLSQISRDKLPDIGSEYEGDADSINVAVEFVLQNF 185
Query: 696 TEMNNFGCGCNIR-PAREKEKR 758
EMN + P REKEKR
Sbjct: 186 IEMNKLWVRMQHQGPVREKEKR 207
>gb|AAO37536.1| putative vacuolar sorting-associated protein, 3'-partial [Oryza
sativa (japonica cultivar-group)]
Length = 198
Score = 328 bits (842), Expect(2) = 2e-89
Identities = 159/184 (86%), Positives = 176/184 (95%), Gaps = 1/184 (0%)
Frame = +3
Query: 159 GTEDEEKFLAAGIAGLQQNSFYMHRALDSNNLRDALKYSAQMLSELRTSKLSPHKYYELY 338
G +DEE++LA GIAG+QQN+FYMHRALDSNNL+DALKYSAQMLSELRTS+LSPHKYY+LY
Sbjct: 6 GADDEERWLAEGIAGVQQNAFYMHRALDSNNLKDALKYSAQMLSELRTSRLSPHKYYDLY 65
Query: 339 MRAFDQLRKLEMFFEEEARRG-CSIIDLYELVQHAGNILPRLYLLCTVGSVYIKSKEAPA 515
MRAFD++RKLEMFF EE RRG CS++DLYELVQHAGN+LPRLYLLCTVGSVYIKSKEAPA
Sbjct: 66 MRAFDEMRKLEMFFREETRRGSCSVVDLYELVQHAGNVLPRLYLLCTVGSVYIKSKEAPA 125
Query: 516 KDVLKDLVEMCRGIQHPVRGLFLRSYLSQVSRDKLPDIGSEYEGDADTVADAVEFVLQNF 695
KDVLKDLVEMCRGIQHP+RGLFLRSYLSQ+SRDKLPDIGSEYEGDAD++ AVEFVLQNF
Sbjct: 126 KDVLKDLVEMCRGIQHPLRGLFLRSYLSQISRDKLPDIGSEYEGDADSINVAVEFVLQNF 185
Query: 696 TEMN 707
EMN
Sbjct: 186 IEMN 189
Score = 23.1 bits (48), Expect(2) = 2e-89
Identities = 8/9 (88%), Positives = 9/9 (99%)
Frame = +2
Query: 707 QLWVRMQHQ 733
+LWVRMQHQ
Sbjct: 190 KLWVRMQHQ 198
>ref|NP_177713.1| vacuolor sorting protein 35, putative; protein id: At1g75850.1
[Arabidopsis thaliana]
Length = 838
Score = 300 bits (768), Expect = 1e-80
Identities = 156/238 (65%), Positives = 183/238 (76%), Gaps = 35/238 (14%)
Frame = +3
Query: 153 LDGTEDEEKFLAAGIAGLQQNSFYMHRALDSNNLRDALKYSAQMLSELRTSKLSPHKYYE 332
L G EDE+K+LA GIAG+Q N+F+MHRALD+NNLR+ LKYSA MLSELRTSKLSP KYY+
Sbjct: 4 LAGVEDEDKWLAEGIAGIQHNAFFMHRALDANNLREVLKYSALMLSELRTSKLSPQKYYD 63
Query: 333 LY----------------------------MRAFDQLRKLEMFFEEEARRGCSIIDLYEL 428
L MRAFDQLR+LE+FF++E+R G ++DLYEL
Sbjct: 64 LCRFHRQRTLNLSIHACVDQFLIIFPSNLDMRAFDQLRQLEIFFKDESRHGLPVVDLYEL 123
Query: 429 VQHAGNILPRLYLLCTVGSVYIKSKEAPAKDVLKDLVEMCRGIQHPVRGLFLRSYLSQVS 608
VQHAGNILPR+YLLCTVGSVYIKSK+AP+KDVLKDLVEMCRG+QHP+RGLFLRSYL+QVS
Sbjct: 124 VQHAGNILPRMYLLCTVGSVYIKSKQAPSKDVLKDLVEMCRGVQHPIRGLFLRSYLAQVS 183
Query: 609 RDKLPDIGSEYEGDADTVADAVEFVLQNFTEMNNF-------GCGCNIRPAREKEKRE 761
RDKLP+IGS+YEGDA+TV DAVEFVLQNFTEMN G G +R +EKE+ E
Sbjct: 184 RDKLPEIGSDYEGDANTVMDAVEFVLQNFTEMNKLWVRIQHQGPG-TVREKQEKERNE 240
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 631,726,678
Number of Sequences: 1393205
Number of extensions: 13287506
Number of successful extensions: 50872
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 45062
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49889
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 36974710344
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)