Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000813A_C01 KMC000813A_c01
(1147 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
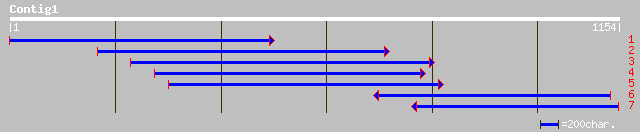
ref|NP_178101.1| unknown protein; protein id: At1g79830.1 [Arabi... 207 3e-52
ref|NP_009045.1| TATA element modulatory factor 1 [Homo sapiens]... 80 8e-14
ref|XP_132740.1| similar to TATA element modulatory factor 1 [Ho... 79 1e-13
emb|CAB97305.2| related to transcription factor TMF [Neurospora ... 70 6e-11
pir||T50985 related to transcription factor TMF [imported] - Neu... 70 6e-11
>ref|NP_178101.1| unknown protein; protein id: At1g79830.1 [Arabidopsis thaliana]
gi|25406610|pir||C96829 unknown protein F19K16.21
[imported] - Arabidopsis thaliana
gi|12324587|gb|AAG52248.1|AC011717_16 unknown protein;
70672-76070 [Arabidopsis thaliana]
Length = 918
Score = 207 bits (526), Expect = 3e-52
Identities = 115/183 (62%), Positives = 137/183 (74%), Gaps = 7/183 (3%)
Frame = -1
Query: 1147 TARVHSA----QTPTTKLNSAFENGNLSRKVSSASSLGSLEESHFLQASLDSSDSYSERR 980
TAR++S+ Q P + NSAFEN D +SE+R
Sbjct: 765 TARINSSAVSEQLPIARQNSAFEN-----------------------------DKFSEKR 795
Query: 979 ---ELSMSPYYMKSMTPSSFEAALRQKEGELASYMSRLASLESIRDSLADELVKLTAQCE 809
E +MSPYYMKS+TPS++EA LRQKEGELASYM+RLAS+ESIRDSLA+ELVK+TA+CE
Sbjct: 796 SMPEATMSPYYMKSITPSAYEATLRQKEGELASYMTRLASMESIRDSLAEELVKMTAECE 855
Query: 808 KLRAEAAVLPGLRSELEALRRRHSAALELMGERDEELEELRADIVDLKEMYREQVNLLVN 629
KLR EA +PG+++ELEALR+RH+AALELMGERDEELEELRADIVDLKEMYREQVN+LVN
Sbjct: 856 KLRGEADRVPGIKAELEALRQRHAAALELMGERDEELEELRADIVDLKEMYREQVNMLVN 915
Query: 628 KIQ 620
KIQ
Sbjct: 916 KIQ 918
>ref|NP_009045.1| TATA element modulatory factor 1 [Homo sapiens]
gi|8134736|sp|P82094|TMF1_HUMAN TATA element modulatory
factor (TMF) gi|423112|pir||A47212 transcription factor
TMF, TATA element modulatory factor - human
gi|5870866|gb|AAD54608.1| TATA element modulatory factor
[Homo sapiens]
Length = 1093
Score = 79.7 bits (195), Expect = 8e-14
Identities = 38/103 (36%), Positives = 67/103 (64%)
Frame = -1
Query: 934 SFEAALRQKEGELASYMSRLASLESIRDSLADELVKLTAQCEKLRAEAAVLPGLRSELEA 755
+ ++ L+ +EGE+ + +LE R +A+ELVKLT Q ++L + +P LR++L
Sbjct: 988 NLQSQLKLREGEITHLQLEIGNLEKTRSIMAEELVKLTNQNDELEEKVKEIPKLRTQLRD 1047
Query: 754 LRRRHSAALELMGERDEELEELRADIVDLKEMYREQVNLLVNK 626
L +R++ L++ GE+ EE EELR D+ D+K MY+ Q++ L+ +
Sbjct: 1048 LDQRYNTILQMYGEKAEEAEELRLDLEDVKNMYKTQIDELLRQ 1090
>ref|XP_132740.1| similar to TATA element modulatory factor 1 [Homo sapiens] [Mus
musculus]
Length = 266
Score = 79.0 bits (193), Expect = 1e-13
Identities = 51/163 (31%), Positives = 89/163 (54%), Gaps = 15/163 (9%)
Frame = -1
Query: 1069 VSSASSLGSLEESHFLQASLDSSDSYSERRELSMSPYYMKSMTPSSFEAA---------- 920
+S +SS+ ++ + LQAS S D E + S P + + +EA
Sbjct: 105 MSRSSSISGVDAAG-LQASFLSQD---ESHDHSFGPMSTSASGSNLYEAVRMGAGSSIIE 160
Query: 919 -----LRQKEGELASYMSRLASLESIRDSLADELVKLTAQCEKLRAEAAVLPGLRSELEA 755
L+ +EGE++ +++LE R +++ELVKLT Q ++L + +P LR +L
Sbjct: 161 NLQSQLKLREGEISHLQLEISNLEKTRSIMSEELVKLTNQNDELEEKVKEIPKLRVQLRD 220
Query: 754 LRRRHSAALELMGERDEELEELRADIVDLKEMYREQVNLLVNK 626
L +R++ L++ GE+ EE EELR D+ D+K MY+ Q++ L+ +
Sbjct: 221 LDQRYNTILQMYGEKAEEAEELRLDLEDVKNMYKTQIDELLRQ 263
>emb|CAB97305.2| related to transcription factor TMF [Neurospora crassa]
gi|28927163|gb|EAA36119.1| hypothetical protein (
(AL389891) related to transcription factor TMF
[Neurospora crassa] )
Length = 900
Score = 70.1 bits (170), Expect = 6e-11
Identities = 48/159 (30%), Positives = 75/159 (46%), Gaps = 14/159 (8%)
Frame = -1
Query: 1078 SRKVSSASSLGSLEESHFLQASLDSSDSYSERRELSMSPY-YMKSMTPSSFEAA------ 920
S + S G E +A L SD+ + + S SP+ ++ M S AA
Sbjct: 737 SSAIHQTSDAGGAPEIQHPRAGLHRSDTGFDSVDSSSSPHNVLQDMVSVSTIAAGPSVQL 796
Query: 919 -------LRQKEGELASYMSRLASLESIRDSLADELVKLTAQCEKLRAEAAVLPGLRSEL 761
+RQ E E + LA + RD E+V L + E + A + L ++
Sbjct: 797 VERMSAKIRQLESEKVTVREELARISKQRDEARAEIVALMGEVENQKKAAERVAELERQV 856
Query: 760 EALRRRHSAALELMGERDEELEELRADIVDLKEMYREQV 644
+ R+ LEL+GE+ EE++EL+AD+ DLK+MYR+ V
Sbjct: 857 AEVNERYETTLELLGEKSEEVDELKADVQDLKDMYRDLV 895
>pir||T50985 related to transcription factor TMF [imported] - Neurospora crassa
Length = 892
Score = 70.1 bits (170), Expect = 6e-11
Identities = 48/159 (30%), Positives = 75/159 (46%), Gaps = 14/159 (8%)
Frame = -1
Query: 1078 SRKVSSASSLGSLEESHFLQASLDSSDSYSERRELSMSPY-YMKSMTPSSFEAA------ 920
S + S G E +A L SD+ + + S SP+ ++ M S AA
Sbjct: 729 SSAIHQTSDAGGAPEIQHPRAGLHRSDTGFDSVDSSSSPHNVLQDMVSVSTIAAGPSVQL 788
Query: 919 -------LRQKEGELASYMSRLASLESIRDSLADELVKLTAQCEKLRAEAAVLPGLRSEL 761
+RQ E E + LA + RD E+V L + E + A + L ++
Sbjct: 789 VERMSAKIRQLESEKVTVREELARISKQRDEARAEIVALMGEVENQKKAAERVAELERQV 848
Query: 760 EALRRRHSAALELMGERDEELEELRADIVDLKEMYREQV 644
+ R+ LEL+GE+ EE++EL+AD+ DLK+MYR+ V
Sbjct: 849 AEVNERYETTLELLGEKSEEVDELKADVQDLKDMYRDLV 887
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 965,859,521
Number of Sequences: 1393205
Number of extensions: 21276319
Number of successful extensions: 80214
Number of sequences better than 10.0: 510
Number of HSP's better than 10.0 without gapping: 61616
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 72716
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 70281887232
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)