Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
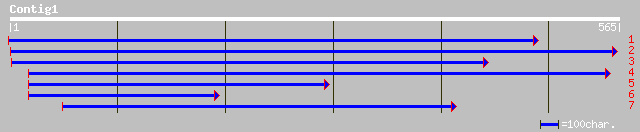
Query= KMC000795A_C01 KMC000795A_c01
(545 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P42347|P3K1_SOYBN PHOSPHATIDYLINOSITOL 3-KINASE, ROOT ISOFORM... 144 9e-34
sp|P42348|P3K2_SOYBN PHOSPHATIDYLINOSITOL 3-KINASE, NODULE ISOFO... 144 9e-34
emb|CAD56881.1| phosphatidylinositol 3-kinase [Medicago truncatula] 142 3e-33
gb|AAN62481.1| phosphatidylinositol 3-kinase [Brassica napus] 140 1e-32
ref|NP_176251.1| phosphatidylinositol 3-kinase, putative; protei... 138 4e-32
>sp|P42347|P3K1_SOYBN PHOSPHATIDYLINOSITOL 3-KINASE, ROOT ISOFORM (PI3-KINASE)
(PTDINS-3-KINASE) (PI3K) (SPI3K-5)
gi|7434345|pir||T07761 phosphatidylinositol 3-kinase -
soybean gi|736339|gb|AAA83995.1| phosphatidylinositol
3-kinase
Length = 814
Score = 144 bits (362), Expect = 9e-34
Identities = 69/72 (95%), Positives = 71/72 (97%)
Frame = -2
Query: 544 LILNLFYLMAGSDIPDIASDPDKGILKLQEKFRLDLDDEASTHFFQDLINESVSALFPQM 365
LILNLFYLMAGS+IPDIASDP+KGILKLQEKFRLDLDDEAS HFFQDLINESVSALFPQM
Sbjct: 743 LILNLFYLMAGSNIPDIASDPEKGILKLQEKFRLDLDDEASIHFFQDLINESVSALFPQM 802
Query: 364 VETIHRWAQYWR 329
VETIHRWAQYWR
Sbjct: 803 VETIHRWAQYWR 814
>sp|P42348|P3K2_SOYBN PHOSPHATIDYLINOSITOL 3-KINASE, NODULE ISOFORM (PI3-KINASE)
(PTDINS-3-KINASE) (PI3K) (SPI3K-1)
gi|7434344|pir||T07745 phosphatidylinositol 3-kinase
PI3K - soybean gi|736337|gb|AAA64468.1|
phosphatidylinositol 3-kinase
Length = 812
Score = 144 bits (362), Expect = 9e-34
Identities = 69/72 (95%), Positives = 71/72 (97%)
Frame = -2
Query: 544 LILNLFYLMAGSDIPDIASDPDKGILKLQEKFRLDLDDEASTHFFQDLINESVSALFPQM 365
LILNLFYLMAGS+IPDIASDP+KGILKLQEKFRLDLDDEAS HFFQDLINESVSALFPQM
Sbjct: 741 LILNLFYLMAGSNIPDIASDPEKGILKLQEKFRLDLDDEASIHFFQDLINESVSALFPQM 800
Query: 364 VETIHRWAQYWR 329
VETIHRWAQYWR
Sbjct: 801 VETIHRWAQYWR 812
>emb|CAD56881.1| phosphatidylinositol 3-kinase [Medicago truncatula]
Length = 808
Score = 142 bits (357), Expect = 3e-33
Identities = 68/72 (94%), Positives = 70/72 (96%)
Frame = -2
Query: 544 LILNLFYLMAGSDIPDIASDPDKGILKLQEKFRLDLDDEASTHFFQDLINESVSALFPQM 365
LILNLFYLMAGS+IPDIASDP+KGILKLQEKFRLDLDDEA HFFQDLINESVSALFPQM
Sbjct: 737 LILNLFYLMAGSNIPDIASDPEKGILKLQEKFRLDLDDEACIHFFQDLINESVSALFPQM 796
Query: 364 VETIHRWAQYWR 329
VETIHRWAQYWR
Sbjct: 797 VETIHRWAQYWR 808
>gb|AAN62481.1| phosphatidylinositol 3-kinase [Brassica napus]
Length = 813
Score = 140 bits (353), Expect = 1e-32
Identities = 67/72 (93%), Positives = 69/72 (95%)
Frame = -2
Query: 544 LILNLFYLMAGSDIPDIASDPDKGILKLQEKFRLDLDDEASTHFFQDLINESVSALFPQM 365
LILNLFYLMAGS IPDIASDP+KGILKLQEKFRLD+DDEA HFFQDLINESVSALFPQM
Sbjct: 742 LILNLFYLMAGSTIPDIASDPEKGILKLQEKFRLDMDDEACIHFFQDLINESVSALFPQM 801
Query: 364 VETIHRWAQYWR 329
VETIHRWAQYWR
Sbjct: 802 VETIHRWAQYWR 813
>ref|NP_176251.1| phosphatidylinositol 3-kinase, putative; protein id: At1g60490.1,
supported by cDNA: gi_555699 [Arabidopsis thaliana]
gi|27735233|sp|P42339|PI3K_ARATH Phosphatidylinositol
3-kinase (PI3-kinase) (PtdIns-3-kinase) (PI3K) (ATVPS34)
gi|25287674|pir||B96630 Phosphatidylinositol 3-kinase
[imported] - Arabidopsis thaliana
gi|2462752|gb|AAB71971.1| Phosphatidylinositol 3-kinase
[Arabidopsis thaliana] gi|24030195|gb|AAN41278.1|
putative phosphatidylinositol 3-kinase [Arabidopsis
thaliana]
Length = 814
Score = 138 bits (348), Expect = 4e-32
Identities = 66/72 (91%), Positives = 69/72 (95%)
Frame = -2
Query: 544 LILNLFYLMAGSDIPDIASDPDKGILKLQEKFRLDLDDEASTHFFQDLINESVSALFPQM 365
LILNLF+LMAGS IPDIASDP+KGILKLQEKFRLD+DDEA HFFQDLINESVSALFPQM
Sbjct: 743 LILNLFHLMAGSTIPDIASDPEKGILKLQEKFRLDMDDEACIHFFQDLINESVSALFPQM 802
Query: 364 VETIHRWAQYWR 329
VETIHRWAQYWR
Sbjct: 803 VETIHRWAQYWR 814
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,294,234
Number of Sequences: 1393205
Number of extensions: 9590683
Number of successful extensions: 19576
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 19210
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19573
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18660035355
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)