Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000790A_C01 KMC000790A_c01
(725 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
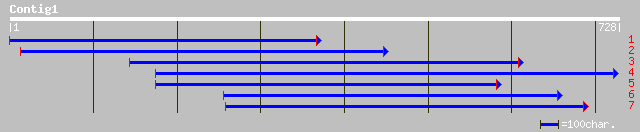
ref|XP_043272.4| similar to KIAA0346 [Homo sapiens] 35 1.1
dbj|BAB19397.1| P0458A05.5 [Oryza sativa (japonica cultivar-group)] 34 1.8
pir||E89453 protein F35H12.3 [imported] - Caenorhabditis elegans 33 5.3
gb|EAA33970.1| predicted protein [Neurospora crassa] 32 6.9
>ref|XP_043272.4| similar to KIAA0346 [Homo sapiens]
Length = 366
Score = 35.0 bits (79), Expect = 1.1
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 3/34 (8%)
Frame = +2
Query: 443 CNHHPRPPP---QRCLAPCLPAVEPAITYNPFHF 535
C+ PRPPP QR +A CLP V P +T F
Sbjct: 48 CSPRPRPPPPQEQRAMAACLPPVPPEVTLERAEF 81
>dbj|BAB19397.1| P0458A05.5 [Oryza sativa (japonica cultivar-group)]
Length = 669
Score = 34.3 bits (77), Expect = 1.8
Identities = 21/45 (46%), Positives = 27/45 (59%), Gaps = 9/45 (20%)
Frame = +2
Query: 440 TCNHHPRPP------PQRCLAPC---LPAVEPAITYNPFHFSTTS 547
TC HP+PP P+R L P LP+ +PA T +PFH +TTS
Sbjct: 167 TC--HPQPPASSNRRPRRPLPPPIPHLPSSQPAWTRDPFHLATTS 209
>pir||E89453 protein F35H12.3 [imported] - Caenorhabditis elegans
Length = 836
Score = 32.7 bits (73), Expect = 5.3
Identities = 30/100 (30%), Positives = 49/100 (49%), Gaps = 9/100 (9%)
Frame = +2
Query: 131 ITLQVTTPATKLKGIIQT------SYYIKRWVKPSSHTEKQRLNHTIARDSSR-SSGLQN 289
+ LQ TP++ KG + T ++ +++K +K RL T S + GL N
Sbjct: 538 LLLQDVTPSSNTKGTVVTLSGSPGTHNDFKYMKSFFEQKKIRLICTNYPGSEFVTGGLHN 597
Query: 290 LRISTNKAYYY*LLMQDHILTKLNRMIITGHSK--HNAMQ 403
+ ++ Y LM+ L +NR+II GHS+ NA+Q
Sbjct: 598 SYTNQDRNSYMKSLMETLELKNVNRLIIMGHSRGGENALQ 637
>gb|EAA33970.1| predicted protein [Neurospora crassa]
Length = 1395
Score = 32.3 bits (72), Expect = 6.9
Identities = 23/89 (25%), Positives = 35/89 (38%), Gaps = 9/89 (10%)
Frame = +1
Query: 346 PHKTKPDDHNRPFKTQRDANTWQEINKKDTIYMQP----PPTAPTSAVLGSLSPRCRACN 513
PH+ P H P Q +++ ++ P PP AP V + SPR
Sbjct: 758 PHQHPPQAHQVPQVPQASQAPQPHVSRAPQVHPTPHLPHPPQAPLPQVTQAQSPRIPPIT 817
Query: 514 HI--QPISF*HHIIY---THKQPLDQNQP 585
H P++ H+ + +QP Q QP
Sbjct: 818 HAPQPPVTHAPHVAHPPQQQQQPQQQQQP 846
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 601,593,820
Number of Sequences: 1393205
Number of extensions: 12967312
Number of successful extensions: 29566
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 28343
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29550
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34062062287
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)