Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000772A_C01 KMC000772A_c01
(739 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
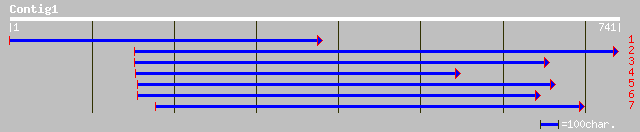
pir||B85062 probable WD-repeat membrane protein [imported] - Ara... 121 9e-27
ref|NP_567275.1| putative WD-repeat membrane protein; protein id... 121 9e-27
gb|AAO51690.1| similar to Arabidopsis thaliana (Mouse-ear cress)... 47 2e-04
ref|NP_705460.1| hypothetical protein [Plasmodium falciparum 3D7... 47 3e-04
gb|EAA09168.1| agCP14169 [Anopheles gambiae str. PEST] 40 0.026
>pir||B85062 probable WD-repeat membrane protein [imported] - Arabidopsis thaliana
gi|4325344|gb|AAD17343.1| similar to beta-transducins
(Pfam: PF00400, Score=71.7, E=1.5e-17, N=6) [Arabidopsis
thaliana] gi|7267253|emb|CAB81036.1| putative WD-repeat
membrane protein [Arabidopsis thaliana]
Length = 931
Score = 121 bits (304), Expect = 9e-27
Identities = 56/83 (67%), Positives = 71/83 (85%)
Frame = -2
Query: 729 RPEFVSIEQLLDYFIYELSCRNNFEFIQAVIRLFLKIHGETIRKQPHLQEKARKLLDVQR 550
RPEF+ I QLLDYFI E+SC+N+FEF+QAV++LFLKIHGETIR P LQEKA+KLL+ Q
Sbjct: 847 RPEFILIGQLLDYFINEVSCKNDFEFMQAVVKLFLKIHGETIRCHPSLQEKAKKLLETQS 906
Query: 549 MVWERVDKLFPSTRCWVAFLSQT 481
+VW++++KLF STRC V FLS +
Sbjct: 907 LVWQKMEKLFQSTRCIVTFLSNS 929
>ref|NP_567275.1| putative WD-repeat membrane protein; protein id: At4g04940.1,
supported by cDNA: gi_16604678, supported by cDNA:
gi_20465602 [Arabidopsis thaliana]
gi|16604679|gb|AAL24132.1| putative WD-repeat membrane
protein [Arabidopsis thaliana] gi|20465603|gb|AAM20284.1|
putative WD-repeat membrane protein [Arabidopsis
thaliana]
Length = 910
Score = 121 bits (304), Expect = 9e-27
Identities = 56/83 (67%), Positives = 71/83 (85%)
Frame = -2
Query: 729 RPEFVSIEQLLDYFIYELSCRNNFEFIQAVIRLFLKIHGETIRKQPHLQEKARKLLDVQR 550
RPEF+ I QLLDYFI E+SC+N+FEF+QAV++LFLKIHGETIR P LQEKA+KLL+ Q
Sbjct: 826 RPEFILIGQLLDYFINEVSCKNDFEFMQAVVKLFLKIHGETIRCHPSLQEKAKKLLETQS 885
Query: 549 MVWERVDKLFPSTRCWVAFLSQT 481
+VW++++KLF STRC V FLS +
Sbjct: 886 LVWQKMEKLFQSTRCIVTFLSNS 908
>gb|AAO51690.1| similar to Arabidopsis thaliana (Mouse-ear cress). Putative WD-repeat
membrane protein [Dictyostelium discoideum]
Length = 1026
Score = 47.4 bits (111), Expect = 2e-04
Identities = 22/81 (27%), Positives = 44/81 (54%)
Frame = -2
Query: 723 EFVSIEQLLDYFIYELSCRNNFEFIQAVIRLFLKIHGETIRKQPHLQEKARKLLDVQRMV 544
++ I + D+ +Y+L +++FIQ+++ + LK+HG + + R+LL V
Sbjct: 944 DYADIHYMFDFIVYQLLKNGDYDFIQSILSVTLKVHGYLL-YDTTFKRDLRQLLLVFNEP 1002
Query: 543 WERVDKLFPSTRCWVAFLSQT 481
W ++ KLF S C + + + T
Sbjct: 1003 WFKMQKLFHSNICMLRYFTNT 1023
>ref|NP_705460.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23615705|emb|CAD52697.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1218
Score = 47.0 bits (110), Expect = 3e-04
Identities = 23/77 (29%), Positives = 44/77 (56%), Gaps = 2/77 (2%)
Frame = -2
Query: 711 IEQLLDYFIYELSCRNNFEFIQAVIRLFLKIHGETI--RKQPHLQEKARKLLDVQRMVWE 538
+E ++++FIY + +N + IQA + +FLK HG+ I K+ L+ + LL + W
Sbjct: 1137 LENMMNFFIYHVKTNDNVDLIQAYLFIFLKAHGKKIMKSKEKKLKNTTKVLLQEIQGCWS 1196
Query: 537 RVDKLFPSTRCWVAFLS 487
++ LF + ++ FL+
Sbjct: 1197 NINFLFENIIFFIKFLT 1213
>gb|EAA09168.1| agCP14169 [Anopheles gambiae str. PEST]
Length = 796
Score = 40.4 bits (93), Expect = 0.026
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 1/56 (1%)
Frame = -2
Query: 687 IYELSCRNNFEFIQAVIRLFLKIHGETIRKQPHLQEKARKLLDVQRMVWERV-DKL 523
+Y L R FE Q+ + LFLK+HG TI + L + + D Q+ W + DKL
Sbjct: 726 VYMLGQRKEFELAQSYLSLFLKLHGRTIVENEQLSKLLPAIEDAQKAEWSALEDKL 781
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 616,699,136
Number of Sequences: 1393205
Number of extensions: 12916789
Number of successful extensions: 25751
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 25114
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25749
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 35188080875
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)