Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
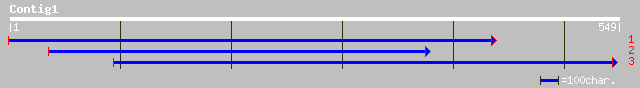
Query= KMC000768A_C01 KMC000768A_c01
(545 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565513.1| RRM-containing protein; protein id: At2g21440.1... 61 8e-09
ref|NP_200811.1| KED - like protein; protein id: At5g60030.1 [Ar... 47 1e-04
ref|NP_494864.2| Protein-tyrosine phosphatase family member [Cae... 45 7e-04
ref|NP_701231.1| hypothetical protein [Plasmodium falciparum 3D7... 43 0.003
ref|NP_702538.1| hypothetical protein [Plasmodium falciparum 3D7... 42 0.004
>ref|NP_565513.1| RRM-containing protein; protein id: At2g21440.1, supported by cDNA:
gi_16604630 [Arabidopsis thaliana]
gi|25412039|pir||C84601 hypothetical protein At2g21440
[imported] - Arabidopsis thaliana
gi|4567275|gb|AAD23688.1| expressed protein [Arabidopsis
thaliana] gi|16604631|gb|AAL24108.1| unknown protein
[Arabidopsis thaliana] gi|27754736|gb|AAO22811.1| unknown
protein [Arabidopsis thaliana]
Length = 1003
Score = 61.2 bits (147), Expect = 8e-09
Identities = 42/115 (36%), Positives = 60/115 (51%), Gaps = 7/115 (6%)
Frame = -2
Query: 541 KNKNAEVSSPKEKPEALPMESKNNQDDQTKSGNKGDVGFRK-----RKVKTKEEQ-EAGK 380
K+ AE P E K Q ++ K D K RK K E++ E +
Sbjct: 888 KDNAAEKKRPIRTQEKPSSNKKGQLMRQKETTEKPDPKISKDLSEPRKRKFGEDRGEENR 947
Query: 379 KVSRKRQKKKK-DSGEEVVDKLDMLIEQYRNKFSQKSPGNGGEKKPTKPLRKWFQ 218
RKR+K+ + G EVVDKLD+LIE+YR+KFSQ S G +K+ + +R+WF+
Sbjct: 948 NGQRKRKKQGQGQGGAEVVDKLDLLIEKYRSKFSQSSAKTGPQKQSSGQVRRWFE 1002
>ref|NP_200811.1| KED - like protein; protein id: At5g60030.1 [Arabidopsis thaliana]
gi|8777342|dbj|BAA96932.1| gene_id:MGO3.1~unknown
protein [Arabidopsis thaliana]
Length = 292
Score = 47.0 bits (110), Expect = 1e-04
Identities = 36/110 (32%), Positives = 58/110 (52%), Gaps = 1/110 (0%)
Frame = -2
Query: 541 KNKNAEVSSPKEKPEALPMESKNNQDDQTKSGNKGDVGFRKRKVKTKEEQEAGKKVSRKR 362
KN + +V KEK E E K+ + + K DV K K K ++EQ +G++ +K
Sbjct: 192 KNNDEDVVDEKEKLED---EQKSAEIKEKKKNKDEDVVDEKEKEKLEDEQRSGER--KKE 246
Query: 361 QKKKKDSGEEVVDKLDMLIEQYRNKFSQKSPGN-GGEKKPTKPLRKWFQI 215
+KKK+ S EE+V E+ ++K +KS G E++ +K RK +I
Sbjct: 247 KKKKRKSDEEIVS------EERKSKKKRKSDEEMGSEERKSKKKRKLKEI 290
Score = 43.1 bits (100), Expect = 0.002
Identities = 30/102 (29%), Positives = 54/102 (52%), Gaps = 6/102 (5%)
Frame = -2
Query: 541 KNKNAEVSSPK--EKPEALPMESKNNQDDQTKSGNKG----DVGFRKRKVKTKEEQEAGK 380
K+K+A+V K EK EA + + + K K DV K K K ++EQ++
Sbjct: 122 KSKDADVVDEKVNEKLEAEQRSEERRERKKEKKKKKNNKDEDVVDEKVKEKLEDEQKSAD 181
Query: 379 KVSRKRQKKKKDSGEEVVDKLDMLIEQYRNKFSQKSPGNGGE 254
+ RK++K KK++ E+VVD+ + L ++ ++ ++ N E
Sbjct: 182 RKERKKKKSKKNNDEDVVDEKEKLEDEQKSAEIKEKKKNKDE 223
Score = 33.9 bits (76), Expect = 1.3
Identities = 24/77 (31%), Positives = 36/77 (46%), Gaps = 4/77 (5%)
Frame = -2
Query: 541 KNKNAEVSSPKEKPEALPMESKNNQDDQTKSGNKGDVGF----RKRKVKTKEEQEAGKKV 374
KNK+ +V KEK + + + + K K D RK K K K ++E G
Sbjct: 219 KNKDEDVVDEKEKEKLEDEQRSGERKKEKKKKRKSDEEIVSEERKSKKKRKSDEEMG--- 275
Query: 373 SRKRQKKKKDSGEEVVD 323
S +R+ KKK +E+ D
Sbjct: 276 SEERKSKKKRKLKEIDD 292
Score = 32.3 bits (72), Expect = 3.8
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Frame = -2
Query: 499 EALPMES---KNNQDDQTKSGNKGDVGFRKRKVKTKEEQEAGKKVSRKRQKKKK--DSGE 335
EA+ +ES + + + K DV K K + EQ + ++ RK++KKKK + E
Sbjct: 103 EAVSVESVYGRERDEKKMKKSKDADVVDEKVNEKLEAEQRSEERRERKKEKKKKKNNKDE 162
Query: 334 EVVDK 320
+VVD+
Sbjct: 163 DVVDE 167
>ref|NP_494864.2| Protein-tyrosine phosphatase family member [Caenorhabditis elegans]
gi|16259241|gb|AAL16312.1|U49830_11 Hypothetical protein
C33F10.8 [Caenorhabditis elegans]
Length = 463
Score = 44.7 bits (104), Expect = 7e-04
Identities = 32/104 (30%), Positives = 50/104 (47%)
Frame = -2
Query: 538 NKNAEVSSPKEKPEALPMESKNNQDDQTKSGNKGDVGFRKRKVKTKEEQEAGKKVSRKRQ 359
+KN E++ P++ + SK Q D K D G RK + +K++ + KKV RK
Sbjct: 2 DKNNELTLPEKTGQDNNQGSKKGQTDSKKK----DQGSRKARGNSKKKNHSSKKVVRKTS 57
Query: 358 KKKKDSGEEVVDKLDMLIEQYRNKFSQKSPGNGGEKKPTKPLRK 227
KKKK++ LI ++ + KSP G KK + L +
Sbjct: 58 KKKKNAASPNA----QLIPVAKSPGNDKSPAGFGRKKAGENLNE 97
>ref|NP_701231.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23496297|gb|AAN35955.1|AE014841_38 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 3468
Score = 42.7 bits (99), Expect = 0.003
Identities = 35/115 (30%), Positives = 53/115 (45%), Gaps = 10/115 (8%)
Frame = -2
Query: 541 KNKNAEVSSPKEKPEALPMESKNNQDDQTKSGNKGDVGFRKRKVKTKEEQEAGKKVSRKR 362
K KN E KEK E E+K ++ + + K + RK K +T+E +E +K RK
Sbjct: 2735 KKKNEE----KEKMENDEKEAKREKEKKEERKEKERIEERKEKERTEERKEKERKEERKE 2790
Query: 361 QKKK-----KDSGEEVVDKL-----DMLIEQYRNKFSQKSPGNGGEKKPTKPLRK 227
++KK K+ EE +KL D E+ + K K EKK ++K
Sbjct: 2791 KEKKEEKNEKEKKEEKNEKLSGKKRDKENEKEKEKKKDKEKKKDNEKKKENEIKK 2845
>ref|NP_702538.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23497724|gb|AAN37262.1|AE014827_5 hypothetical protein
[Plasmodium falciparum 3D7]
Length = 2558
Score = 42.4 bits (98), Expect = 0.004
Identities = 26/97 (26%), Positives = 45/97 (45%)
Frame = -2
Query: 544 HKNKNAEVSSPKEKPEALPMESKNNQDDQTKSGNKGDVGFRKRKVKTKEEQEAGKKVSRK 365
++N N SS +E+ E P ++++ DD S N +K K+ +E+EA K
Sbjct: 2454 NRNYNNGKSSNEEENEGTPADNEDENDDSNNSNNSNQE--QKEKILENDEEEATDDNEEK 2511
Query: 364 RQKKKKDSGEEVVDKLDMLIEQYRNKFSQKSPGNGGE 254
+++K D EE E+Y N ++K N +
Sbjct: 2512 NEEEKNDDNEEE--------EEYENNDNEKYNKNNSD 2540
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 467,087,241
Number of Sequences: 1393205
Number of extensions: 10130035
Number of successful extensions: 45427
Number of sequences better than 10.0: 587
Number of HSP's better than 10.0 without gapping: 37598
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43254
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18660035355
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)