Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000764A_C01 KMC000764A_c01
(549 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
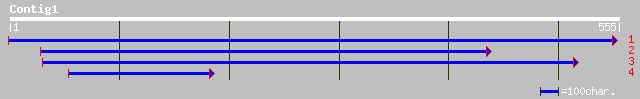
ref|NP_181015.1| unknown protein; protein id: At2g34680.1 [Arabi... 159 3e-38
pir||T51336 auxin-induced protein AIR9 [imported] - Arabidopsis ... 156 2e-37
ref|NP_325967.1| unknown; predicted coding region [Mycoplasma pu... 33 2.3
ref|NP_070878.1| 2-ketoisovalerate ferredoxin oxidoreductase, su... 32 5.0
sp|P40122|CAP_CHLVR ADENYLYL CYCLASE-ASSOCIATED PROTEIN (CAP) gi... 31 8.6
>ref|NP_181015.1| unknown protein; protein id: At2g34680.1 [Arabidopsis thaliana]
gi|7487522|pir||T01367 hypothetical protein At2g34680
[imported] - Arabidopsis thaliana
gi|3132477|gb|AAC16266.1| unknown protein [Arabidopsis
thaliana]
Length = 1680
Score = 159 bits (401), Expect = 3e-38
Identities = 77/98 (78%), Positives = 87/98 (88%)
Frame = -3
Query: 535 GTYERRILEINRKRLKVLKPATKTSFPNTEIRGSYAPPFHVELFRNDQHRLKIVVDSENE 356
G ERR+LE+NRKR+KV+KP +KTSF TE+RGSY PPFHVE FRNDQ RL+IVVDSENE
Sbjct: 1583 GNLERRMLEMNRKRIKVVKPGSKTSFATTEVRGSYGPPFHVETFRNDQRRLRIVVDSENE 1642
Query: 355 ADLMVHSRHLRDVIVLVIRGLAQRFNSTSLNSLLKIET 242
D++V SRHLRDVIVLVIRG AQRFNSTSLNSLLKI+T
Sbjct: 1643 VDIVVQSRHLRDVIVLVIRGFAQRFNSTSLNSLLKIDT 1680
>pir||T51336 auxin-induced protein AIR9 [imported] - Arabidopsis thaliana
(fragment) gi|3695021|gb|AAC62612.1| hypothetical
protein [Arabidopsis thaliana]
Length = 234
Score = 156 bits (394), Expect = 2e-37
Identities = 76/96 (79%), Positives = 85/96 (88%)
Frame = -3
Query: 535 GTYERRILEINRKRLKVLKPATKTSFPNTEIRGSYAPPFHVELFRNDQHRLKIVVDSENE 356
G ERR+LE+NRKR+KV+KP +KTSF TE+RGSY PPFHVE FRNDQ RL+IVVDSENE
Sbjct: 134 GNLERRMLEMNRKRIKVVKPGSKTSFATTEVRGSYGPPFHVETFRNDQRRLRIVVDSENE 193
Query: 355 ADLMVHSRHLRDVIVLVIRGLAQRFNSTSLNSLLKI 248
D++V SRHLRDVIVLVIRG AQRFNSTSLNSLLKI
Sbjct: 194 VDIVVQSRHLRDVIVLVIRGFAQRFNSTSLNSLLKI 229
>ref|NP_325967.1| unknown; predicted coding region [Mycoplasma pulmonis]
gi|25376254|pir||H90528 hypothetical protein MYPU_1360
[imported] - Mycoplasma pulmonis (strain UAB CTIP)
gi|14089549|emb|CAC13309.1| unknown; predicted coding
region [Mycoplasma pulmonis]
Length = 148
Score = 33.1 bits (74), Expect = 2.3
Identities = 12/25 (48%), Positives = 20/25 (80%)
Frame = -3
Query: 385 LKIVVDSENEADLMVHSRHLRDVIV 311
+K ++ ENE DL+V+S HLRD+++
Sbjct: 27 MKKEIEKENEFDLIVYSEHLRDIVI 51
>ref|NP_070878.1| 2-ketoisovalerate ferredoxin oxidoreductase, subunit delta (vorD)
[Archaeoglobus fulgidus] gi|7430791|pir||E69506 probable
2-oxoisovalerate-ferredoxin oxidoreductase (EC 1.2.7.-)
delta chain AF2054 [similarity] - Archaeoglobus fulgidus
gi|2648489|gb|AAB89211.1| 2-ketoisovalerate ferredoxin
oxidoreductase, subunit delta (vorD) [Archaeoglobus
fulgidus DSM 4304]
Length = 82
Score = 32.0 bits (71), Expect = 5.0
Identities = 19/51 (37%), Positives = 21/51 (40%), Gaps = 1/51 (1%)
Frame = -2
Query: 206 TLYSCISSGKKRFLRPVTSHFACPECK-CCCYVERSAINALIIAGIDY*IC 57
T S +G R LRPV C CK C Y I + A IDY C
Sbjct: 11 TAGSAGETGSWRILRPVVDEKKCSGCKECFYYCPEGVIQVDVFARIDYRFC 61
>sp|P40122|CAP_CHLVR ADENYLYL CYCLASE-ASSOCIATED PROTEIN (CAP) gi|630480|pir||S47091
cyclase-associated protein - Chlorohydra viridissima
gi|498781|emb|CAA56104.1| cyclase associated protein
[Chlorohydra viridissima]
Length = 481
Score = 31.2 bits (69), Expect = 8.6
Identities = 13/26 (50%), Positives = 18/26 (69%)
Frame = +3
Query: 9 FLMQNLKW*LSNQCEHTDLVINACND 86
F +QN KW + NQ +T+L I+ CND
Sbjct: 326 FQLQNKKWVIENQDGNTNLEISECND 351
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 419,111,105
Number of Sequences: 1393205
Number of extensions: 8094875
Number of successful extensions: 14990
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 14685
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 14988
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)