Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000759A_C01 KMC000759A_c01
(514 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
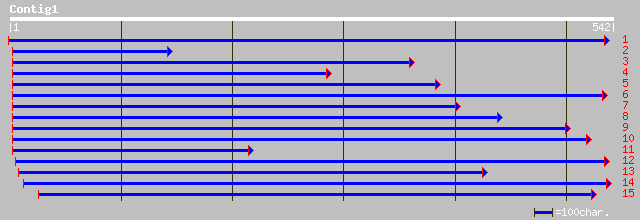
emb|CAB76909.1| lipoxygenase [Cicer arietinum] 214 5e-55
pir||T06454 probable lipoxygenase (EC 1.13.11.12) - garden pea g... 211 5e-54
emb|CAA55318.1| lipoxygenase [Pisum sativum] 209 1e-53
sp|P14856|LOX2_PEA Seed lipoxygenase-2 gi|100055|pir||S07075 lip... 209 1e-53
pir||T06827 lipoxygenase (EC 1.13.11.12) - garden pea gi|2459611... 209 2e-53
>emb|CAB76909.1| lipoxygenase [Cicer arietinum]
Length = 540
Score = 214 bits (545), Expect = 5e-55
Identities = 105/127 (82%), Positives = 116/127 (90%)
Frame = -3
Query: 512 RRLIPEKGTPEYDEMVKNPQKAYLRTITPKYQALVDLSVIEILSRHASDEIYLGERENPN 333
RR IPEKGTPEYDEMVK+PQKAYLRTITPKYQ LVDLSVIEILSRHASDE+YLGER++
Sbjct: 414 RRFIPEKGTPEYDEMVKSPQKAYLRTITPKYQTLVDLSVIEILSRHASDEVYLGERDSKF 473
Query: 332 WTSDSKALQAFKKFGTKLQEIEAKITERNKDSSLKNRIGPVELPYTLLVPSSEAGLTFRG 153
WTSDS+A+QAF+KFG+KL EIE KI ERNKDSSLKNR GP++LPY LL+ SSE GLTFRG
Sbjct: 474 WTSDSRAVQAFQKFGSKLTEIEGKIIERNKDSSLKNRYGPIQLPYNLLLRSSEEGLTFRG 533
Query: 152 IPNSISI 132
IPNSISI
Sbjct: 534 IPNSISI 540
>pir||T06454 probable lipoxygenase (EC 1.13.11.12) - garden pea
gi|2598612|emb|CAA75609.1| lipoxygenase [Pisum sativum]
Length = 866
Score = 211 bits (536), Expect = 5e-54
Identities = 104/127 (81%), Positives = 112/127 (87%)
Frame = -3
Query: 512 RRLIPEKGTPEYDEMVKNPQKAYLRTITPKYQALVDLSVIEILSRHASDEIYLGERENPN 333
RRLIPEKGTP YDEMVKNPQKAYLRTITPK+Q L+DLSVIEILSRHASDEIYLGER++
Sbjct: 740 RRLIPEKGTPHYDEMVKNPQKAYLRTITPKFQTLIDLSVIEILSRHASDEIYLGERDSKF 799
Query: 332 WTSDSKALQAFKKFGTKLQEIEAKITERNKDSSLKNRIGPVELPYTLLVPSSEAGLTFRG 153
WTSDS+ALQAFKKFG KL +IE I ERN DSSLKNR GPV+LPYT+L PS E GL FRG
Sbjct: 800 WTSDSRALQAFKKFGNKLAKIEGTIKERNNDSSLKNRYGPVQLPYTILQPSGEDGLAFRG 859
Query: 152 IPNSISI 132
IPNSISI
Sbjct: 860 IPNSISI 866
>emb|CAA55318.1| lipoxygenase [Pisum sativum]
Length = 863
Score = 209 bits (533), Expect = 1e-53
Identities = 101/127 (79%), Positives = 116/127 (90%)
Frame = -3
Query: 512 RRLIPEKGTPEYDEMVKNPQKAYLRTITPKYQALVDLSVIEILSRHASDEIYLGERENPN 333
RRL+PE+GT EYDEMVK+ QKAYLRTITPK+Q L+DLSVIEILSRHASDE+YLG+RENP+
Sbjct: 737 RRLLPEEGTAEYDEMVKSSQKAYLRTITPKFQTLIDLSVIEILSRHASDEVYLGQRENPH 796
Query: 332 WTSDSKALQAFKKFGTKLQEIEAKITERNKDSSLKNRIGPVELPYTLLVPSSEAGLTFRG 153
WTSDSKALQAF+KFG KL EIEAK+T +N D SL +R+GPV+LPYTLL PSS+ GLTFRG
Sbjct: 797 WTSDSKALQAFQKFGNKLAEIEAKLTNKNNDPSLYHRVGPVQLPYTLLHPSSKEGLTFRG 856
Query: 152 IPNSISI 132
IPNSISI
Sbjct: 857 IPNSISI 863
>sp|P14856|LOX2_PEA Seed lipoxygenase-2 gi|100055|pir||S07075 lipoxygenase (EC
1.13.11.12) 2 [similarity] - garden pea
gi|20802|emb|CAA34906.1| lipoxygenase (AA 1-864) [Pisum
sativum]
Length = 864
Score = 209 bits (533), Expect = 1e-53
Identities = 101/127 (79%), Positives = 116/127 (90%)
Frame = -3
Query: 512 RRLIPEKGTPEYDEMVKNPQKAYLRTITPKYQALVDLSVIEILSRHASDEIYLGERENPN 333
RRL+PE+GT EYDEMVK+ QKAYLRTITPK+Q L+DLSVIEILSRHASDE+YLG+RENP+
Sbjct: 738 RRLLPEEGTAEYDEMVKSSQKAYLRTITPKFQTLIDLSVIEILSRHASDEVYLGQRENPH 797
Query: 332 WTSDSKALQAFKKFGTKLQEIEAKITERNKDSSLKNRIGPVELPYTLLVPSSEAGLTFRG 153
WTSDSKALQAF+KFG KL EIEAK+T +N D SL +R+GPV+LPYTLL PSS+ GLTFRG
Sbjct: 798 WTSDSKALQAFQKFGNKLAEIEAKLTNKNNDPSLYHRVGPVQLPYTLLHPSSKEGLTFRG 857
Query: 152 IPNSISI 132
IPNSISI
Sbjct: 858 IPNSISI 864
>pir||T06827 lipoxygenase (EC 1.13.11.12) - garden pea gi|2459611|gb|AAB71759.1|
lipoxygenase [Pisum sativum]
Length = 868
Score = 209 bits (531), Expect = 2e-53
Identities = 102/127 (80%), Positives = 112/127 (87%)
Frame = -3
Query: 512 RRLIPEKGTPEYDEMVKNPQKAYLRTITPKYQALVDLSVIEILSRHASDEIYLGERENPN 333
RR IPEKGTPEYDEMVK+PQKAYLRTITPKYQ LVDLSVIEILSRHASDE+YLGER+N N
Sbjct: 742 RRFIPEKGTPEYDEMVKSPQKAYLRTITPKYQTLVDLSVIEILSRHASDEVYLGERDNKN 801
Query: 332 WTSDSKALQAFKKFGTKLQEIEAKITERNKDSSLKNRIGPVELPYTLLVPSSEAGLTFRG 153
WTSDS+A+QAF KFGTKL EIE KI RN + L+NR GPV+LPYTLL+ SSE GLTFR
Sbjct: 802 WTSDSRAVQAFAKFGTKLTEIEGKIHSRNNEPGLRNRYGPVQLPYTLLLRSSEEGLTFRR 861
Query: 152 IPNSISI 132
IPNS+SI
Sbjct: 862 IPNSVSI 868
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 416,934,312
Number of Sequences: 1393205
Number of extensions: 8265030
Number of successful extensions: 22087
Number of sequences better than 10.0: 147
Number of HSP's better than 10.0 without gapping: 21585
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22034
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 16232377112
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)