Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
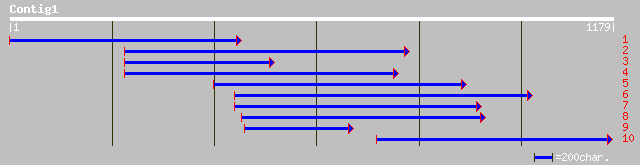
Query= KMC000757A_C01 KMC000757A_c01
(1171 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB08225.1| gene_id:MUF9.5~gi|5174629~similar to unknown pro... 158 2e-37
ref|NP_200850.1| putative protein; protein id: At5g60420.1 [Arab... 158 2e-37
gb|EAA04501.1| agCP3526 [Anopheles gambiae str. PEST] 36 1.0
sp|P41319|ANFC_SQUAC C-type natriuretic peptide precursor gi|539... 35 2.3
emb|CAA16808.1| /prediction=(method:''genefinder'', version:''08... 35 3.0
>dbj|BAB08225.1| gene_id:MUF9.5~gi|5174629~similar to unknown protein [Arabidopsis
thaliana] gi|22654962|gb|AAM98074.1| AT5g60420/muf9_70
[Arabidopsis thaliana] gi|28416515|gb|AAO42788.1|
AT5g60420/muf9_70 [Arabidopsis thaliana]
Length = 873
Score = 158 bits (399), Expect = 2e-37
Identities = 81/151 (53%), Positives = 107/151 (70%)
Frame = -2
Query: 1170 DLNGGLVDHPLAFAGDDPSLQIFLPPRPAESSVPNELRDRXDVSHGVCTEDWISLSLGGG 991
+ N GLVD+PLAF DDPSLQIFLP +P ++S + +++ D+S+G+ +EDWISL LG
Sbjct: 736 EANDGLVDNPLAFGRDDPSLQIFLPTKP-DASAQSGFKNQADMSNGLRSEDWISLRLGDS 794
Query: 990 AAGSNGNASAQNGLNSRGNASIQNGLNSRHQVPAMDDDTHTLADTASLLLGMNDARSDKA 811
A+G++G+ + NG+NS HQ+ + T +TASLLLGMND+R DKA
Sbjct: 795 ASGNHGDPATTNGINSS------------HQMSTREGSMDTTTETASLLLGMNDSRQDKA 842
Query: 810 SRPRSGSPFSFPRQKRSVRPRLYLSIDSDSE 718
+ RS +PFSFPRQKRSVRPR+YLSIDSDSE
Sbjct: 843 KKQRSDNPFSFPRQKRSVRPRMYLSIDSDSE 873
>ref|NP_200850.1| putative protein; protein id: At5g60420.1 [Arabidopsis thaliana]
Length = 509
Score = 158 bits (399), Expect = 2e-37
Identities = 81/151 (53%), Positives = 107/151 (70%)
Frame = -2
Query: 1170 DLNGGLVDHPLAFAGDDPSLQIFLPPRPAESSVPNELRDRXDVSHGVCTEDWISLSLGGG 991
+ N GLVD+PLAF DDPSLQIFLP +P ++S + +++ D+S+G+ +EDWISL LG
Sbjct: 372 EANDGLVDNPLAFGRDDPSLQIFLPTKP-DASAQSGFKNQADMSNGLRSEDWISLRLGDS 430
Query: 990 AAGSNGNASAQNGLNSRGNASIQNGLNSRHQVPAMDDDTHTLADTASLLLGMNDARSDKA 811
A+G++G+ + NG+NS HQ+ + T +TASLLLGMND+R DKA
Sbjct: 431 ASGNHGDPATTNGINSS------------HQMSTREGSMDTTTETASLLLGMNDSRQDKA 478
Query: 810 SRPRSGSPFSFPRQKRSVRPRLYLSIDSDSE 718
+ RS +PFSFPRQKRSVRPR+YLSIDSDSE
Sbjct: 479 KKQRSDNPFSFPRQKRSVRPRMYLSIDSDSE 509
>gb|EAA04501.1| agCP3526 [Anopheles gambiae str. PEST]
Length = 576
Score = 36.2 bits (82), Expect = 1.0
Identities = 33/127 (25%), Positives = 51/127 (39%), Gaps = 10/127 (7%)
Frame = -2
Query: 1101 LPPRPAESSVPNELRDRXDVSHGVCTEDWISLSLGGGAAGSNGNASAQNGLNSRGNASIQ 922
L P P SV N G+ S GGG G GNA++ + + + GN S
Sbjct: 288 LTPMPFHDSVHNICETTTGSGSGIMVMSATSTMAGGGGGG-GGNATSSSPITTNGNGSTH 346
Query: 921 --NGLNSRHQVPAMDDDTHTLADT--------ASLLLGMNDARSDKASRPRSGSPFSFPR 772
+ ++S + +T+++ + SLL +N+ A R RS SP PR
Sbjct: 347 RYSTVSSSSNPGGAEQNTNSVGKSIAGGGGPELSLLTKLNNG---AAERSRSNSPNVVPR 403
Query: 771 QKRSVRP 751
+ P
Sbjct: 404 PEAGPTP 410
>sp|P41319|ANFC_SQUAC C-type natriuretic peptide precursor gi|539462|pir||A61244
natriuretic peptide type C precursor - spiny dogfish
gi|556804|emb|CAA42608.1| prepro atrial natriuretic
factor [Squalus acanthias] gi|228343|prf||1803178A C-type
natriuretic peptide
Length = 135
Score = 35.0 bits (79), Expect = 2.3
Identities = 23/69 (33%), Positives = 32/69 (46%)
Frame = +2
Query: 965 EALPLLPAAPPPRLRDIQSSVQTPWDTSXRSLNSFGTDDSAGLGGRKI*SEGSSPAKAKG 1144
EA + PAA P L QS ++ PWD R + A L R + ++P + KG
Sbjct: 51 EAQEISPAASLPDLNTDQSDLELPWDRESREIGGRSFRQEA-LLARLLQDLSNNPLRFKG 109
Query: 1145 WSTKPPFKS 1171
S K P +S
Sbjct: 110 RSKKGPSRS 118
>emb|CAA16808.1| /prediction=(method:''genefinder'', version:''084'',
score:''77.81'')~/prediction=(method:''genscan'',
version:''1.0'', score:''153.79'')~/motif=(desc:''ATP
synthase alpha and beta subunits signature'',
dbase:''PROSITE'', acc:''PS00152'', method:''ppsearch'')
[Drosophila melanogaster]
Length = 449
Score = 34.7 bits (78), Expect = 3.0
Identities = 20/50 (40%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Frame = -2
Query: 1035 GVCTEDWISLSLGGG----AAGSNGNASAQNGLNSRGNASIQNGLNSRHQ 898
GV D S ++GGG AAG+ G ++A NS N+++ LNS+HQ
Sbjct: 385 GVVASDATSGAIGGGGGAGAAGAAGASAAAAHSNSNSNSNLNLNLNSQHQ 434
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 972,518,468
Number of Sequences: 1393205
Number of extensions: 21087024
Number of successful extensions: 58037
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 51626
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 57600
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 72478196208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)