Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000754A_C01 KMC000754A_c01
(953 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
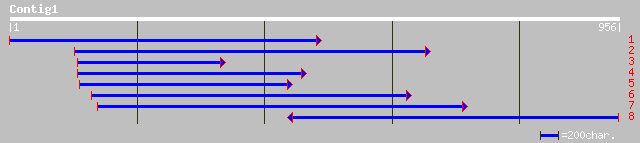
gb|AAH32196.1| Unknown (protein for MGC:38244) [Mus musculus] gi... 449 e-125
gb|AAL33589.1| methionine synthase [Zea mays] 438 e-122
sp|P93263|METE_MESCR 5-methyltetrahydropteroyltriglutamate--homo... 436 e-121
gb|AAF74983.1|AF082893_1 methionine synthase [Solanum tuberosum] 436 e-120
sp|Q42699|METE_CATRO 5-methyltetrahydropteroyltriglutamate--homo... 428 e-119
>gb|AAH32196.1| Unknown (protein for MGC:38244) [Mus musculus]
gi|22028279|gb|AAH34830.1| Unknown (protein for
MGC:28753) [Mus musculus]
Length = 498
Score = 449 bits (1155), Expect = e-125
Identities = 217/249 (87%), Positives = 237/249 (95%)
Frame = +3
Query: 177 MASHIVGYPRMGPKRELKFALESFWDGKTSADDLKKVSADLRASIWKQMAGAGIKYIPSN 356
MASHIVGYPRMGPKRELKFALESFWDGK+SADDLKKVSADLR+SIWKQM+ AGIKYIPSN
Sbjct: 1 MASHIVGYPRMGPKRELKFALESFWDGKSSADDLKKVSADLRSSIWKQMSDAGIKYIPSN 60
Query: 357 TFSYYDGVLDATATLGAVPPRYGWTGGEIGFDTYFSMARGNATVPAMEMTKWFDTNYHFI 536
TF+YYD VLD TA LGAVPPRYGWTGGEI FD YFSMARGNATVPAMEMTKWFDTNYH+I
Sbjct: 61 TFAYYDQVLDTTAMLGAVPPRYGWTGGEIEFDVYFSMARGNATVPAMEMTKWFDTNYHYI 120
Query: 537 VPELGPEVKFAYSSHKAVDEYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKSFSLLSL 716
VPELGP+VKF+Y+SHKAV+EYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKSF+LLSL
Sbjct: 121 VPELGPDVKFSYASHKAVNEYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKSFNLLSL 180
Query: 717 LPKVLAVYKEVIADLKAAGASWIQFDEPTLVLDLDSHKLAAFTEAYSELASSLSGLNVLV 896
LPK+LA+YKEV+ +LKAAGAS IQFDEP LV+DLDSHKL AFT+AY+EL S+LSGLNV+V
Sbjct: 181 LPKILAIYKEVVTELKAAGASCIQFDEPLLVMDLDSHKLQAFTDAYAELESTLSGLNVIV 240
Query: 897 ETYFADILP 923
ETYFAD+ P
Sbjct: 241 ETYFADVTP 249
>gb|AAL33589.1| methionine synthase [Zea mays]
Length = 766
Score = 438 bits (1127), Expect = e-122
Identities = 216/247 (87%), Positives = 229/247 (92%)
Frame = +3
Query: 177 MASHIVGYPRMGPKRELKFALESFWDGKTSADDLKKVSADLRASIWKQMAGAGIKYIPSN 356
MASHIVGYPRMGPKRELKFALESFWDGK++A+DL+KV+ DLRASIWKQMA AGIKYIPSN
Sbjct: 1 MASHIVGYPRMGPKRELKFALESFWDGKSTAEDLEKVATDLRASIWKQMADAGIKYIPSN 60
Query: 357 TFSYYDGVLDATATLGAVPPRYGWTGGEIGFDTYFSMARGNATVPAMEMTKWFDTNYHFI 536
TFSYYD VLD TA LGAVP RY WTGGEIGFDTYFSMARGNATVPAMEMTKWFDTNYHFI
Sbjct: 61 TFSYYDQVLDTTAMLGAVPERYSWTGGEIGFDTYFSMARGNATVPAMEMTKWFDTNYHFI 120
Query: 537 VPELGPEVKFAYSSHKAVDEYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKSFSLLSL 716
VPELGP KF+Y+SHKAV+EYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEK F LLSL
Sbjct: 121 VPELGPNTKFSYASHKAVNEYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKGFPLLSL 180
Query: 717 LPKVLAVYKEVIADLKAAGASWIQFDEPTLVLDLDSHKLAAFTEAYSELASSLSGLNVLV 896
L +L VYKEVIA+LKAAGASWIQFDEPTLVLDLDS KLAAF+ AY+EL S LSGLNVLV
Sbjct: 181 LSSILPVYKEVIAELKAAGASWIQFDEPTLVLDLDSDKLAAFSAAYAELESVLSGLNVLV 240
Query: 897 ETYFADI 917
ETYFAD+
Sbjct: 241 ETYFADV 247
>sp|P93263|METE_MESCR 5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (Vitamin-B12-independent methionine
synthase isozyme) (Cobalamin-independent methionine
synthase isozyme) gi|7433427|pir||T12575
5-methyltetrahydropteroyltriglutamate-homocysteine
S-methyltransferase (EC 2.1.1.14) - common ice plant
gi|1814403|gb|AAB41896.1| methionine synthase
[Mesembryanthemum crystallinum]
Length = 765
Score = 436 bits (1120), Expect = e-121
Identities = 211/247 (85%), Positives = 231/247 (93%)
Frame = +3
Query: 177 MASHIVGYPRMGPKRELKFALESFWDGKTSADDLKKVSADLRASIWKQMAGAGIKYIPSN 356
MASHIVGYPRMGPKRELKFALESFWDGK++A+DLKKVSADLR+SIWKQMA AGIKYIPSN
Sbjct: 1 MASHIVGYPRMGPKRELKFALESFWDGKSTAEDLKKVSADLRSSIWKQMADAGIKYIPSN 60
Query: 357 TFSYYDGVLDATATLGAVPPRYGWTGGEIGFDTYFSMARGNATVPAMEMTKWFDTNYHFI 536
TFSYYD VLD TA LGAVPPRYGWTGGEI FD YFSMARGNA+VPAMEMTKWFDTNYHFI
Sbjct: 61 TFSYYDQVLDTTAMLGAVPPRYGWTGGEIEFDVYFSMARGNASVPAMEMTKWFDTNYHFI 120
Query: 537 VPELGPEVKFAYSSHKAVDEYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKSFSLLSL 716
VPELGPEV F+Y+SHKAV EYKEAKALGVDTVPVLVGPVSYLLLSK AKGV+KSF LLSL
Sbjct: 121 VPELGPEVNFSYASHKAVLEYKEAKALGVDTVPVLVGPVSYLLLSKQAKGVDKSFDLLSL 180
Query: 717 LPKVLAVYKEVIADLKAAGASWIQFDEPTLVLDLDSHKLAAFTEAYSELASSLSGLNVLV 896
LPK+L +YKEV+A+LK AGASWIQFDEP LV+DL+SHKL AF+ AY++L S+LSGLNV+V
Sbjct: 181 LPKILPIYKEVVAELKEAGASWIQFDEPLLVMDLESHKLQAFSAAYADLESTLSGLNVVV 240
Query: 897 ETYFADI 917
ETYFAD+
Sbjct: 241 ETYFADV 247
>gb|AAF74983.1|AF082893_1 methionine synthase [Solanum tuberosum]
Length = 765
Score = 436 bits (1121), Expect(2) = e-120
Identities = 213/247 (86%), Positives = 231/247 (93%)
Frame = +3
Query: 177 MASHIVGYPRMGPKRELKFALESFWDGKTSADDLKKVSADLRASIWKQMAGAGIKYIPSN 356
MASH+VGYPRMGPKRELKFALESFWDGK+SA+DLKKVSADLR+SIWKQM+ AGIKYIPSN
Sbjct: 1 MASHVVGYPRMGPKRELKFALESFWDGKSSAEDLKKVSADLRSSIWKQMSDAGIKYIPSN 60
Query: 357 TFSYYDGVLDATATLGAVPPRYGWTGGEIGFDTYFSMARGNATVPAMEMTKWFDTNYHFI 536
TFSYYD VLD TA LGAVP RY WTGGEI F TYFSMARGNA+VPAMEMTKWFDTNYHFI
Sbjct: 61 TFSYYDQVLDTTAMLGAVPSRYNWTGGEIEFGTYFSMARGNASVPAMEMTKWFDTNYHFI 120
Query: 537 VPELGPEVKFAYSSHKAVDEYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKSFSLLSL 716
VPELGP+V F+Y+SHKAV+EYKEAKA GVDTVPVLVGPVSYLLLSKPAKGVEKSF LLSL
Sbjct: 121 VPELGPDVNFSYASHKAVNEYKEAKAQGVDTVPVLVGPVSYLLLSKPAKGVEKSFPLLSL 180
Query: 717 LPKVLAVYKEVIADLKAAGASWIQFDEPTLVLDLDSHKLAAFTEAYSELASSLSGLNVLV 896
L K+L +YKEVIA+LKAAGASWIQ DEPTLVLDL+SHKL AFT+AY++L SSLSGLNVLV
Sbjct: 181 LDKILPIYKEVIAELKAAGASWIQLDEPTLVLDLESHKLEAFTKAYADLESSLSGLNVLV 240
Query: 897 ETYFADI 917
ETYFAD+
Sbjct: 241 ETYFADV 247
Score = 20.8 bits (42), Expect(2) = e-120
Identities = 8/10 (80%), Positives = 9/10 (90%)
Frame = +2
Query: 917 PXEAFKTLTS 946
P EAFKTLT+
Sbjct: 248 PAEAFKTLTA 257
>sp|Q42699|METE_CATRO 5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (Vitamin-B12-independent methionine
synthase isozyme) (Cobalamin-independent methionine
synthase isozyme) gi|1362086|pir||S57636
5-methyltetrahydropteroyltriglutamate-homocysteine
S-methyltransferase (EC 2.1.1.14) - Madagascar
periwinkle gi|886471|emb|CAA58474.1| methionine synthase
[Catharanthus roseus]
Length = 765
Score = 428 bits (1101), Expect = e-119
Identities = 209/247 (84%), Positives = 230/247 (92%)
Frame = +3
Query: 177 MASHIVGYPRMGPKRELKFALESFWDGKTSADDLKKVSADLRASIWKQMAGAGIKYIPSN 356
MASHIVGYPRMGPKRELKFALESFWD K+SA+DL+KV+ADLR+SIWKQMA AGIKYIPSN
Sbjct: 1 MASHIVGYPRMGPKRELKFALESFWDKKSSAEDLQKVAADLRSSIWKQMADAGIKYIPSN 60
Query: 357 TFSYYDGVLDATATLGAVPPRYGWTGGEIGFDTYFSMARGNATVPAMEMTKWFDTNYHFI 536
TFSYYD VLD LGAVPPRY + GGEIGFDTYFSMARGNA+VPAMEMTKWFDTNYH+I
Sbjct: 61 TFSYYDQVLDTATMLGAVPPRYNFAGGEIGFDTYFSMARGNASVPAMEMTKWFDTNYHYI 120
Query: 537 VPELGPEVKFAYSSHKAVDEYKEAKALGVDTVPVLVGPVSYLLLSKPAKGVEKSFSLLSL 716
VPELGPEV F+Y+SHKAV+EYKEAK LGVDTVPVLVGPV++LLLSKPAKGVEK+F LLSL
Sbjct: 121 VPELGPEVNFSYASHKAVNEYKEAKELGVDTVPVLVGPVTFLLLSKPAKGVEKTFPLLSL 180
Query: 717 LPKVLAVYKEVIADLKAAGASWIQFDEPTLVLDLDSHKLAAFTEAYSELASSLSGLNVLV 896
L K+L VYKEVI +LKAAGASWIQFDEPTLVLDL+SH+L AFT+AYSEL S+LSGLNV+V
Sbjct: 181 LDKILPVYKEVIGELKAAGASWIQFDEPTLVLDLESHQLEAFTKAYSELESTLSGLNVIV 240
Query: 897 ETYFADI 917
ETYFADI
Sbjct: 241 ETYFADI 247
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 868,506,946
Number of Sequences: 1393205
Number of extensions: 21212204
Number of successful extensions: 70762
Number of sequences better than 10.0: 152
Number of HSP's better than 10.0 without gapping: 65043
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 70404
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 53801056208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)