Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000695A_C01 KMC000695A_c01
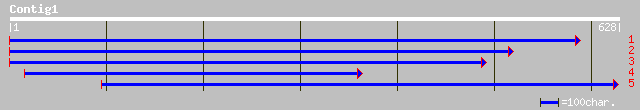
(628 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_197170.1| putative protein; protein id: At5g16680.1 [Arab... 49 7e-05
ref|NP_193211.1| hypothetical protein; protein id: At4g14750.1 [... 44 0.002
gb|AAO73267.1| hypothetical protein [Oryza sativa (japonica cult... 43 0.003
gb|AAL86322.1| unknown protein [Arabidopsis thaliana] 42 0.007
emb|CAD40069.1| OSJNBa0085C10.21 [Oryza sativa (japonica cultiva... 34 0.008
>ref|NP_197170.1| putative protein; protein id: At5g16680.1 [Arabidopsis thaliana]
gi|11357809|pir||T51500 hypothetical protein F5E19_20 -
Arabidopsis thaliana gi|9755720|emb|CAC01832.1| putative
protein [Arabidopsis thaliana]
Length = 1280
Score = 48.5 bits (114), Expect = 7e-05
Identities = 39/95 (41%), Positives = 49/95 (51%), Gaps = 11/95 (11%)
Frame = -1
Query: 574 IPNLELALGGETKLPPPPPPPPPAPKGMLPFLV---GAVERKNNPADS---LADEQED-- 419
+PNLELALG E A G+LPFL + E+ NN + ADE+E+
Sbjct: 1195 VPNLELALGAEETTE--------ATMGLLPFLSRSSNSGEQSNNSMNKEKQKADEEEEDD 1246
Query: 418 ---AASLSLSLSFPSSNKEHKKVDGNHVNTPFLLF 323
AASLSLSLSFP + + +VNTP LF
Sbjct: 1247 AEVAASLSLSLSFPGTEER------KNVNTPLFLF 1275
>ref|NP_193211.1| hypothetical protein; protein id: At4g14750.1 [Arabidopsis
thaliana] gi|7485089|pir||D71410 hypothetical protein -
Arabidopsis thaliana gi|2244832|emb|CAB10254.1|
hypothetical protein [Arabidopsis thaliana]
gi|7268181|emb|CAB78517.1| hypothetical protein
[Arabidopsis thaliana]
Length = 314
Score = 43.9 bits (102), Expect = 0.002
Identities = 22/61 (36%), Positives = 37/61 (60%), Gaps = 3/61 (4%)
Frame = -1
Query: 532 PPPPPPPPPAPKGMLPFLVGAVERKNNPADSL-ADEQEDAASLSLSLSFPS--SNKEHKK 362
PPPPPPPPP P PF+V V+ ++ ++ A+E E+ A++ + + S N+E+ K
Sbjct: 67 PPPPPPPPPPPPLQQPFVVEIVDNEDEQIKNVSAEEIEEFAAIKIQACYRSHLENEENIK 126
Query: 361 V 359
+
Sbjct: 127 I 127
>gb|AAO73267.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 286
Score = 43.1 bits (100), Expect = 0.003
Identities = 30/73 (41%), Positives = 36/73 (49%), Gaps = 2/73 (2%)
Frame = +3
Query: 384 DGNDRERERDAASSCSSAKLSAGLFFLSTAPTRKGSMPLGAGGGGGGGG-GSFVSPPNAS 560
DG DRER R+ SS + A T G+ LGAGGGGG GG P+ S
Sbjct: 190 DGGDRERRRELESSEEEGRREA--------ETDGGAAALGAGGGGGSGGRKGMTGGPHPS 241
Query: 561 SR-FGIPSRNRSA 596
+R G P+R R A
Sbjct: 242 ARVVGGPARERGA 254
>gb|AAL86322.1| unknown protein [Arabidopsis thaliana]
Length = 409
Score = 42.0 bits (97), Expect = 0.007
Identities = 19/51 (37%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Frame = -1
Query: 532 PPPPPPPPPAPKGMLPFLVGAVERKNNPADSL-ADEQEDAASLSLSLSFPS 383
PPPPPPPPP P PF+V V+ ++ ++ A+E E+ A++ + + S
Sbjct: 89 PPPPPPPPPPPPLQQPFVVEIVDNEDEQIKNVSAEEIEEFAAIKIQACYRS 139
>emb|CAD40069.1| OSJNBa0085C10.21 [Oryza sativa (japonica cultivar-group)]
Length = 1639
Score = 34.3 bits (77), Expect = 1.4
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = -1
Query: 532 PPPPPPPPPAPKGMLPFLVGAVERKNN 452
PPPPPPPPP P P L + ++ N
Sbjct: 247 PPPPPPPPPPPPDTNPILTQILAQQAN 273
Score = 31.2 bits (69), Expect(2) = 0.008
Identities = 11/15 (73%), Positives = 11/15 (73%)
Frame = -1
Query: 550 GGETKLPPPPPPPPP 506
GG PPPPPPPPP
Sbjct: 244 GGPPPPPPPPPPPPP 258
Score = 29.6 bits (65), Expect(2) = 0.008
Identities = 19/63 (30%), Positives = 25/63 (39%), Gaps = 3/63 (4%)
Frame = -1
Query: 526 PPPPPPPAPKGMLPFL---VGAVERKNNPADSLADEQEDAASLSLSLSFPSSNKEHKKVD 356
PPPPPPP + FL NNP D+L + ++ L N E K +
Sbjct: 291 PPPPPPPQHSKLAEFLHIRPPTFSSSNNPVDAL----DCLHAVGKKLDTVQCNDEEKVIF 346
Query: 355 GNH 347
H
Sbjct: 347 ATH 349
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 576,852,693
Number of Sequences: 1393205
Number of extensions: 14727770
Number of successful extensions: 271733
Number of sequences better than 10.0: 1464
Number of HSP's better than 10.0 without gapping: 89297
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 187753
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25586195130
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)