Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000675A_C01 KMC000675A_c01
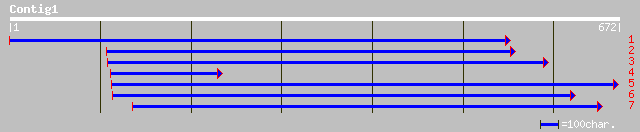
(663 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_567022.1| AP3-complex beta-3A adaptin subunit-like protei... 127 2e-28
gb|AAL24128.1| putative AP3-complex beta-3A adaptin subunit [Ara... 127 2e-28
pir||T47687 adaptor protein/ adaptin-like - Arabidopsis thaliana... 127 2e-28
dbj|BAB63694.1| putative adaptor protein/ adaptin-like [Oryza sa... 107 2e-22
gb|ZP_00094163.1| hypothetical protein [Novosphingobium aromatic... 40 0.028
>ref|NP_567022.1| AP3-complex beta-3A adaptin subunit-like protein; protein id:
At3g55480.1, supported by cDNA: gi_16604670 [Arabidopsis
thaliana]
Length = 987
Score = 127 bits (318), Expect = 2e-28
Identities = 58/97 (59%), Positives = 82/97 (83%)
Frame = -2
Query: 653 EDKFLVICETLAIKMLSHANLSIVSVDMPVASHLDDASGLCLRFCSEILSHSMPCLITVT 474
+DKFL ICE++ +K+LS++NL +VSVD+PVA+ L+DA+GL LRF S+ILS +P LIT+T
Sbjct: 886 KDKFLSICESITLKVLSNSNLHLVSVDLPVANSLEDATGLRLRFSSKILSSEIPLLITIT 945
Query: 473 VEGKCSDPLTASVKVHCEDPVFGLNFLNRVVHFLVEP 363
VEGKC++ L +VK++CE+ VFGLN LNR+ +F+VEP
Sbjct: 946 VEGKCTEVLNLTVKINCEETVFGLNLLNRIANFMVEP 982
>gb|AAL24128.1| putative AP3-complex beta-3A adaptin subunit [Arabidopsis thaliana]
Length = 987
Score = 127 bits (318), Expect = 2e-28
Identities = 58/97 (59%), Positives = 82/97 (83%)
Frame = -2
Query: 653 EDKFLVICETLAIKMLSHANLSIVSVDMPVASHLDDASGLCLRFCSEILSHSMPCLITVT 474
+DKFL ICE++ +K+LS++NL +VSVD+PVA+ L+DA+GL LRF S+ILS +P LIT+T
Sbjct: 886 KDKFLSICESITLKVLSNSNLHLVSVDLPVANSLEDATGLRLRFSSKILSSEIPLLITIT 945
Query: 473 VEGKCSDPLTASVKVHCEDPVFGLNFLNRVVHFLVEP 363
VEGKC++ L +VK++CE+ VFGLN LNR+ +F+VEP
Sbjct: 946 VEGKCTEVLNLTVKINCEETVFGLNLLNRIANFMVEP 982
>pir||T47687 adaptor protein/ adaptin-like - Arabidopsis thaliana (fragment)
gi|7076791|emb|CAB75906.1| adaptor protein/ adaptin-like
[Arabidopsis thaliana]
Length = 1123
Score = 127 bits (318), Expect = 2e-28
Identities = 58/97 (59%), Positives = 82/97 (83%)
Frame = -2
Query: 653 EDKFLVICETLAIKMLSHANLSIVSVDMPVASHLDDASGLCLRFCSEILSHSMPCLITVT 474
+DKFL ICE++ +K+LS++NL +VSVD+PVA+ L+DA+GL LRF S+ILS +P LIT+T
Sbjct: 1022 KDKFLSICESITLKVLSNSNLHLVSVDLPVANSLEDATGLRLRFSSKILSSEIPLLITIT 1081
Query: 473 VEGKCSDPLTASVKVHCEDPVFGLNFLNRVVHFLVEP 363
VEGKC++ L +VK++CE+ VFGLN LNR+ +F+VEP
Sbjct: 1082 VEGKCTEVLNLTVKINCEETVFGLNLLNRIANFMVEP 1118
>dbj|BAB63694.1| putative adaptor protein/ adaptin-like [Oryza sativa (japonica
cultivar-group)] gi|20160932|dbj|BAB89868.1| putative
adaptor protein/ adaptin-like [Oryza sativa (japonica
cultivar-group)]
Length = 1036
Score = 107 bits (266), Expect = 2e-22
Identities = 54/90 (60%), Positives = 67/90 (74%)
Frame = -2
Query: 650 DKFLVICETLAIKMLSHANLSIVSVDMPVASHLDDASGLCLRFCSEILSHSMPCLITVTV 471
DK L+I ++LA K+LS+AN +VS+DMPV ++DASGLC RF SEILS S PCLIT+
Sbjct: 943 DKNLLIAQSLASKILSNANFHLVSMDMPVTFSIEDASGLCWRFSSEILSTSNPCLITILA 1002
Query: 470 EGKCSDPLTASVKVHCEDPVFGLNFLNRVV 381
EG S+PL + KV+ ED FGLN LNRVV
Sbjct: 1003 EGHISEPLDLTAKVNSEDTAFGLNLLNRVV 1032
>gb|ZP_00094163.1| hypothetical protein [Novosphingobium aromaticivorans]
Length = 488
Score = 40.0 bits (92), Expect = 0.028
Identities = 23/58 (39%), Positives = 28/58 (47%)
Frame = -2
Query: 257 CIFFLLVRYCFFFFFLFFYC*KERKGEREMNDCCLFFFG*FNYPDSFFFLSFFFFFGL 84
C FF +CFFFFFLF C+FF F++ FFF+ FF FF L
Sbjct: 5 CRFFFF--FCFFFFFLF----------------CVFFILFFSFSVFFFFIFFFCFFSL 44
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 566,441,819
Number of Sequences: 1393205
Number of extensions: 13088370
Number of successful extensions: 62558
Number of sequences better than 10.0: 147
Number of HSP's better than 10.0 without gapping: 44032
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 54244
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28572683052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)