Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000668A_C01 KMC000668A_c01
(584 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
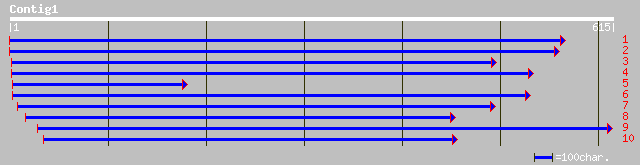
dbj|BAA93022.1| ESTs C74776(E51022),C26123(C116681) correspond t... 114 1e-24
ref|NP_174475.1| G-protein, putative; protein id: At1g31930.1 [A... 111 6e-24
gb|AAL34942.1|AC079037_15 Putative G-protein [Oryza sativa] 96 3e-19
pir||T01135 probable GTP-binding protein (extra large) [imported... 63 3e-09
ref|NP_565553.1| putative GTP-binding protein (extra large); pro... 63 3e-09
>dbj|BAA93022.1| ESTs C74776(E51022),C26123(C116681) correspond to a region of the
predicted gene.~Similar to Arabidopsis thaliana cultivar
Landsberg extra-large G-protein (AF060942) [Oryza sativa
(japonica cultivar-group)]
Length = 867
Score = 114 bits (284), Expect = 1e-24
Identities = 58/91 (63%), Positives = 72/91 (78%), Gaps = 1/91 (1%)
Frame = -2
Query: 583 RPHHNSHALAQQAYYYVAVRFKELYHSLTSQKLFVGQTRARDRASVDEAFKYIREVIKWE 404
R HHN+ +LA QA+YYVA++FKELY + T +KLFV Q RARDR +VDEAFK+IREV+KWE
Sbjct: 779 RTHHNNQSLAHQAFYYVAMKFKELYAACTDRKLFVWQARARDRLTVDEAFKFIREVLKWE 838
Query: 403 DEKDDDVYEINLEESYYS-TEMSSSPFVRQE 314
DEKD Y +ES+YS TE+SSS +RQE
Sbjct: 839 DEKDGGGY--YPDESFYSTTELSSSRLIRQE 867
>ref|NP_174475.1| G-protein, putative; protein id: At1g31930.1 [Arabidopsis thaliana]
gi|25403136|pir||E86443 probable G-protein alpha subunit
[imported] - Arabidopsis thaliana
gi|12321289|gb|AAG50710.1|AC079041_3 G-protein, putative
[Arabidopsis thaliana]
gi|12321467|gb|AAG50792.1|AC074309_9 G-protein alpha
subunit, putative [Arabidopsis thaliana]
Length = 848
Score = 111 bits (278), Expect = 6e-24
Identities = 52/90 (57%), Positives = 71/90 (78%)
Frame = -2
Query: 583 RPHHNSHALAQQAYYYVAVRFKELYHSLTSQKLFVGQTRARDRASVDEAFKYIREVIKWE 404
R ++N +LA QAY+YVA++FK LY S+T QKLFV Q RARDRA+VDE FKY+REV+KW+
Sbjct: 759 RTNNNVQSLAYQAYFYVAMKFKLLYFSITGQKLFVWQARARDRANVDEGFKYVREVLKWD 818
Query: 403 DEKDDDVYEINLEESYYSTEMSSSPFVRQE 314
+EK++ E+S+YST+MSSSP+ +E
Sbjct: 819 EEKEESYLNGGGEDSFYSTDMSSSPYRPEE 848
>gb|AAL34942.1|AC079037_15 Putative G-protein [Oryza sativa]
Length = 583
Score = 96.3 bits (238), Expect = 3e-19
Identities = 47/87 (54%), Positives = 66/87 (75%), Gaps = 2/87 (2%)
Frame = -2
Query: 583 RPHHNSH-ALAQQAYYYVAVRFKELYHSLTS-QKLFVGQTRARDRASVDEAFKYIREVIK 410
RPHH S +LA AYYYVAV+FK+LY S+ +KLFV QT+A +R +VD+AF+YIREV++
Sbjct: 496 RPHHTSQTSLASHAYYYVAVKFKDLYSSVADGRKLFVFQTKALERRTVDDAFRYIREVLR 555
Query: 409 WEDEKDDDVYEINLEESYYSTEMSSSP 329
W+D K+ D + +ES YS +M++SP
Sbjct: 556 WDDVKNSDAGYCSADESSYSVDMTTSP 582
>pir||T01135 probable GTP-binding protein (extra large) [imported] - Arabidopsis
thaliana
Length = 901
Score = 62.8 bits (151), Expect = 3e-09
Identities = 32/81 (39%), Positives = 50/81 (61%)
Frame = -2
Query: 577 HHNSHALAQQAYYYVAVRFKELYHSLTSQKLFVGQTRARDRASVDEAFKYIREVIKWEDE 398
++ + L Q A++++AV+FK Y SLT +KLFV +++ D SVD + K E++KW +E
Sbjct: 821 NNGNPTLGQLAFHFMAVKFKRFYSSLTGKKLFVSSSKSLDPNSVDSSLKLAMEILKWSEE 880
Query: 397 KDDDVYEINLEESYYSTEMSS 335
+ + E S YSTE SS
Sbjct: 881 RTNICMS---EYSMYSTEPSS 898
>ref|NP_565553.1| putative GTP-binding protein (extra large); protein id: At2g23460.1,
supported by cDNA: gi_15451169, supported by cDNA:
gi_3201679 [Arabidopsis thaliana] gi|11357318|pir||T51593
GTP-binding regulatory protein extra-large [validated] -
Arabidopsis thaliana gi|3201680|gb|AAC19352.1|
extra-large G-protein [Arabidopsis thaliana]
gi|20197006|gb|AAC23761.2| putative GTP-binding protein
(extra large) [Arabidopsis thaliana]
Length = 888
Score = 62.8 bits (151), Expect = 3e-09
Identities = 32/81 (39%), Positives = 50/81 (61%)
Frame = -2
Query: 577 HHNSHALAQQAYYYVAVRFKELYHSLTSQKLFVGQTRARDRASVDEAFKYIREVIKWEDE 398
++ + L Q A++++AV+FK Y SLT +KLFV +++ D SVD + K E++KW +E
Sbjct: 808 NNGNPTLGQLAFHFMAVKFKRFYSSLTGKKLFVSSSKSLDPNSVDSSLKLAMEILKWSEE 867
Query: 397 KDDDVYEINLEESYYSTEMSS 335
+ + E S YSTE SS
Sbjct: 868 RTNICMS---EYSMYSTEPSS 885
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 450,941,156
Number of Sequences: 1393205
Number of extensions: 8682382
Number of successful extensions: 25601
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 24389
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25521
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)