Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
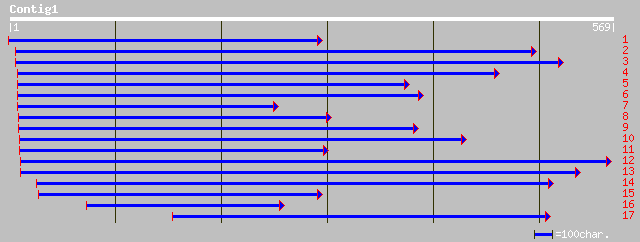
Query= KMC000667A_C01 KMC000667A_c01
(566 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T06779 clathrin heavy chain - soybean gi|1335862|gb|AAC4929... 99 1e-27
ref|NP_187724.1| putative clathrin heavy chain; protein id: At3g... 100 4e-27
gb|AAM19776.1| AT3g08530/T8G24_1 [Arabidopsis thaliana] gi|21928... 100 1e-20
ref|NP_187466.2| clathrin heavy chain, putative; protein id: At3... 100 1e-20
gb|AAG50828.1|AC074395_2 clathrin heavy chain, putative [Arabido... 100 1e-20
>pir||T06779 clathrin heavy chain - soybean gi|1335862|gb|AAC49294.1| clathrin
heavy chain
Length = 1700
Score = 99.0 bits (245), Expect(2) = 1e-27
Identities = 47/60 (78%), Positives = 53/60 (88%)
Frame = -3
Query: 564 KVDELVKDKIEAQIEEKAKEKEEKDVVSQQNMYAQLLPLALPAPPMPGMGGGYGSSPSNG 385
KVDELVKDKIEAQ + KAKE+EEK+V++QQNMYAQLLPLALPAPPMPGMGGG+ P G
Sbjct: 1623 KVDELVKDKIEAQNQVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGFAPPPPMG 1682
Score = 45.8 bits (107), Expect(2) = 1e-27
Identities = 19/22 (86%), Positives = 20/22 (90%), Gaps = 2/22 (9%)
Frame = -1
Query: 401 PPPPMGGLGMP--PPYGMPPMG 342
PPPPMGGLGMP PP+GMPPMG
Sbjct: 1677 PPPPMGGLGMPPMPPFGMPPMG 1698
>ref|NP_187724.1| putative clathrin heavy chain; protein id: At3g11130.1 [Arabidopsis
thaliana] gi|6016683|gb|AAF01510.1|AC009991_6 putative
clathrin heavy chain [Arabidopsis thaliana]
gi|12321871|gb|AAG50967.1|AC073395_9 clathrin heavy
chain, putative; 28833-19741 [Arabidopsis thaliana]
Length = 1705
Score = 100 bits (249), Expect(2) = 4e-27
Identities = 50/61 (81%), Positives = 54/61 (87%), Gaps = 1/61 (1%)
Frame = -3
Query: 564 KVDELVKDKIEAQIEEKAKEKEEKDVVSQQNMYAQLLPLALPAPPMPGM-GGGYGSSPSN 388
KVDEL+KDK+EAQ E KAKE+EEKDV+SQQNMYAQLLPLALPAPPMPGM GGGYG P
Sbjct: 1623 KVDELIKDKLEAQKEVKAKEQEEKDVMSQQNMYAQLLPLALPAPPMPGMGGGGYGPPPQM 1682
Query: 387 G 385
G
Sbjct: 1683 G 1683
Score = 42.4 bits (98), Expect(2) = 4e-27
Identities = 20/28 (71%), Positives = 21/28 (74%), Gaps = 6/28 (21%)
Frame = -1
Query: 404 GPPPPMGGL----GMPP--PYGMPPMGG 339
GPPP MGG+ GMPP PYGMPPMGG
Sbjct: 1677 GPPPQMGGMPGMSGMPPMPPYGMPPMGG 1704
>gb|AAM19776.1| AT3g08530/T8G24_1 [Arabidopsis thaliana] gi|21928019|gb|AAM78038.1|
AT3g08530/T8G24_1 [Arabidopsis thaliana]
Length = 694
Score = 100 bits (249), Expect = 1e-20
Identities = 50/70 (71%), Positives = 56/70 (79%), Gaps = 2/70 (2%)
Frame = -3
Query: 564 KVDELVKDKIEAQIEEKAKEKEEKDVVSQQNMYAQLLPLALPAPPMPGM--GGGYGSSPS 391
KVDEL+KDK+EAQ E KAKE+EEKDV+SQQNMYAQ+LPLALPAPPMPGM GGGYG P
Sbjct: 614 KVDELIKDKLEAQKEVKAKEQEEKDVISQQNMYAQMLPLALPAPPMPGMGGGGGYGPPPQ 673
Query: 390 NGWTRDAPSL 361
G P +
Sbjct: 674 MGGMPGMPPM 683
Score = 43.5 bits (101), Expect = 0.002
Identities = 20/25 (80%), Positives = 21/25 (84%), Gaps = 3/25 (12%)
Frame = -1
Query: 404 GPPPPMGGL-GMPP--PYGMPPMGG 339
GPPP MGG+ GMPP PYGMPPMGG
Sbjct: 669 GPPPQMGGMPGMPPMPPYGMPPMGG 693
>ref|NP_187466.2| clathrin heavy chain, putative; protein id: At3g08530.1 [Arabidopsis
thaliana]
Length = 1674
Score = 100 bits (249), Expect = 1e-20
Identities = 50/70 (71%), Positives = 56/70 (79%), Gaps = 2/70 (2%)
Frame = -3
Query: 564 KVDELVKDKIEAQIEEKAKEKEEKDVVSQQNMYAQLLPLALPAPPMPGM--GGGYGSSPS 391
KVDEL+KDK+EAQ E KAKE+EEKDV+SQQNMYAQ+LPLALPAPPMPGM GGGYG P
Sbjct: 1594 KVDELIKDKLEAQKEVKAKEQEEKDVISQQNMYAQMLPLALPAPPMPGMGGGGGYGPPPQ 1653
Query: 390 NGWTRDAPSL 361
G P +
Sbjct: 1654 MGGMPGMPPM 1663
Score = 43.5 bits (101), Expect = 0.002
Identities = 20/25 (80%), Positives = 21/25 (84%), Gaps = 3/25 (12%)
Frame = -1
Query: 404 GPPPPMGGL-GMPP--PYGMPPMGG 339
GPPP MGG+ GMPP PYGMPPMGG
Sbjct: 1649 GPPPQMGGMPGMPPMPPYGMPPMGG 1673
>gb|AAG50828.1|AC074395_2 clathrin heavy chain, putative [Arabidopsis thaliana]
Length = 1516
Score = 100 bits (249), Expect = 1e-20
Identities = 50/70 (71%), Positives = 56/70 (79%), Gaps = 2/70 (2%)
Frame = -3
Query: 564 KVDELVKDKIEAQIEEKAKEKEEKDVVSQQNMYAQLLPLALPAPPMPGM--GGGYGSSPS 391
KVDEL+KDK+EAQ E KAKE+EEKDV+SQQNMYAQ+LPLALPAPPMPGM GGGYG P
Sbjct: 1436 KVDELIKDKLEAQKEVKAKEQEEKDVISQQNMYAQMLPLALPAPPMPGMGGGGGYGPPPQ 1495
Query: 390 NGWTRDAPSL 361
G P +
Sbjct: 1496 MGGMPGMPPM 1505
Score = 43.5 bits (101), Expect = 0.002
Identities = 20/25 (80%), Positives = 21/25 (84%), Gaps = 3/25 (12%)
Frame = -1
Query: 404 GPPPPMGGL-GMPP--PYGMPPMGG 339
GPPP MGG+ GMPP PYGMPPMGG
Sbjct: 1491 GPPPQMGGMPGMPPMPPYGMPPMGG 1515
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 533,822,315
Number of Sequences: 1393205
Number of extensions: 13972913
Number of successful extensions: 71798
Number of sequences better than 10.0: 180
Number of HSP's better than 10.0 without gapping: 47513
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 69392
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)