Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000663A_C01 KMC000663A_c01
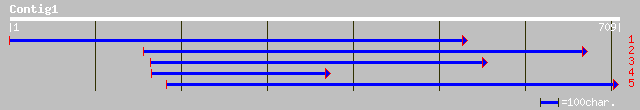
(709 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_174240.1| unknown protein; protein id: At1g29470.1 [Arabi... 179 3e-44
gb|AAM78114.1| AT5g64030/MBM17_13 [Arabidopsis thaliana] gi|2776... 174 1e-42
ref|NP_201208.1| ankyrin-like protein; protein id: At5g64030.1 [... 174 1e-42
dbj|BAB90379.1| ankyrin-like protein [Oryza sativa (japonica cul... 169 3e-41
ref|NP_180977.1| unknown protein; protein id: At2g34300.1 [Arabi... 168 8e-41
>ref|NP_174240.1| unknown protein; protein id: At1g29470.1 [Arabidopsis thaliana]
gi|25403070|pir||E86417 unknown protein, 55790-52851
[imported] - Arabidopsis thaliana
gi|12323540|gb|AAG51752.1|AC068667_31 unknown protein;
55790-52851 [Arabidopsis thaliana]
Length = 768
Score = 179 bits (454), Expect = 3e-44
Identities = 82/114 (71%), Positives = 98/114 (85%)
Frame = -3
Query: 707 IYERGLFXIYHDWCESFSTYPRSYDLLHADSLFSKLKERCNIVAVIAEVDRMLRPQGYLI 528
IYERGLF IYHDWCESFSTYPR+YDLLHAD LFS LK+RCN+V V+AEVDR+LRPQG I
Sbjct: 655 IYERGLFGIYHDWCESFSTYPRTYDLLHADHLFSSLKKRCNLVGVMAEVDRILRPQGTFI 714
Query: 527 IRDNVETIGEIESMTKSLHWDIQFTYSKLGDGLLCIQKTLWRPTEVVTIMSAIA 366
+RD++ETIGEIE M KS+ W+++ T+SK G+GLL +QK+ WRPTE TI SAIA
Sbjct: 715 VRDDMETIGEIEKMVKSMKWNVRMTHSKDGEGLLSVQKSWWRPTEAETIQSAIA 768
>gb|AAM78114.1| AT5g64030/MBM17_13 [Arabidopsis thaliana] gi|27764914|gb|AAO23578.1|
At5g64030/MBM17_13 [Arabidopsis thaliana]
Length = 829
Score = 174 bits (441), Expect = 1e-42
Identities = 78/113 (69%), Positives = 97/113 (85%)
Frame = -3
Query: 707 IYERGLFXIYHDWCESFSTYPRSYDLLHADSLFSKLKERCNIVAVIAEVDRMLRPQGYLI 528
IYERGLF IYHDWCESFSTYPRSYDLLHAD LFSKLK+RCN+ AVIAEVDR+LRP+G LI
Sbjct: 716 IYERGLFGIYHDWCESFSTYPRSYDLLHADHLFSKLKQRCNLTAVIAEVDRVLRPEGKLI 775
Query: 527 IRDNVETIGEIESMTKSLHWDIQFTYSKLGDGLLCIQKTLWRPTEVVTIMSAI 369
+RD+ ETI ++E M K++ W+++ TYSK +GLL +QK++WRP+EV T+ AI
Sbjct: 776 VRDDAETIQQVEGMVKAMKWEVRMTYSKEKEGLLSVQKSIWRPSEVETLTYAI 828
>ref|NP_201208.1| ankyrin-like protein; protein id: At5g64030.1 [Arabidopsis thaliana]
Length = 786
Score = 174 bits (441), Expect = 1e-42
Identities = 78/113 (69%), Positives = 97/113 (85%)
Frame = -3
Query: 707 IYERGLFXIYHDWCESFSTYPRSYDLLHADSLFSKLKERCNIVAVIAEVDRMLRPQGYLI 528
IYERGLF IYHDWCESFSTYPRSYDLLHAD LFSKLK+RCN+ AVIAEVDR+LRP+G LI
Sbjct: 673 IYERGLFGIYHDWCESFSTYPRSYDLLHADHLFSKLKQRCNLTAVIAEVDRVLRPEGKLI 732
Query: 527 IRDNVETIGEIESMTKSLHWDIQFTYSKLGDGLLCIQKTLWRPTEVVTIMSAI 369
+RD+ ETI ++E M K++ W+++ TYSK +GLL +QK++WRP+EV T+ AI
Sbjct: 733 VRDDAETIQQVEGMVKAMKWEVRMTYSKEKEGLLSVQKSIWRPSEVETLTYAI 785
>dbj|BAB90379.1| ankyrin-like protein [Oryza sativa (japonica cultivar-group)]
Length = 806
Score = 169 bits (428), Expect = 3e-41
Identities = 73/110 (66%), Positives = 92/110 (83%)
Frame = -3
Query: 707 IYERGLFXIYHDWCESFSTYPRSYDLLHADSLFSKLKERCNIVAVIAEVDRMLRPQGYLI 528
IYERGLF +YHDWCESFSTYPR+YDLLHAD LFSKLK+RC ++ V AEVDR+LRP+G LI
Sbjct: 697 IYERGLFGMYHDWCESFSTYPRTYDLLHADHLFSKLKKRCKLLPVFAEVDRILRPEGKLI 756
Query: 527 IRDNVETIGEIESMTKSLHWDIQFTYSKLGDGLLCIQKTLWRPTEVVTIM 378
+RDN ETI E++ M KSL W+++ TY+K +GLLC+QK++WRP E+ M
Sbjct: 757 VRDNAETINELQGMVKSLQWEVRMTYTKGNEGLLCVQKSMWRPKEIEASM 806
>ref|NP_180977.1| unknown protein; protein id: At2g34300.1 [Arabidopsis thaliana]
gi|7485462|pir||T02318 hypothetical protein At2g34300
[imported] - Arabidopsis thaliana
gi|3337361|gb|AAC27406.1| unknown protein [Arabidopsis
thaliana]
Length = 770
Score = 168 bits (425), Expect = 8e-41
Identities = 76/114 (66%), Positives = 95/114 (82%)
Frame = -3
Query: 707 IYERGLFXIYHDWCESFSTYPRSYDLLHADSLFSKLKERCNIVAVIAEVDRMLRPQGYLI 528
IYERGLF IYHDWCESF+TYPR+YDLLHAD LFS L++RCN+V+V+AE+DR+LRPQG I
Sbjct: 657 IYERGLFGIYHDWCESFNTYPRTYDLLHADHLFSTLRKRCNLVSVMAEIDRILRPQGTFI 716
Query: 527 IRDNVETIGEIESMTKSLHWDIQFTYSKLGDGLLCIQKTLWRPTEVVTIMSAIA 366
IRD++ET+GE+E M KS+ W ++ T SK +GLL I+K+ WRP E TI SAIA
Sbjct: 717 IRDDMETLGEVEKMVKSMKWKVKMTQSKDNEGLLSIEKSWWRPEETETIKSAIA 770
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 572,111,216
Number of Sequences: 1393205
Number of extensions: 12202599
Number of successful extensions: 62059
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 29531
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56198
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32373034405
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)