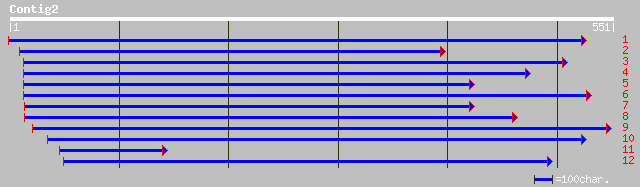
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000655A_C02 KMC000655A_c02
(549 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB11155.1| contains similarity to unknown protein~gene_id:M... 150 7e-36
ref|NP_196314.2| putative protein; protein id: At5g06970.1, supp... 150 7e-36
gb|AAK14418.1|AC087851_10 unknown protein [Oryza sativa] 117 1e-25
ref|NP_192904.1| hypothetical protein; protein id: At4g11670.1 [... 80 2e-14
ref|NP_180900.1| unknown protein; protein id: At2g33420.1 [Arabi... 66 2e-10
>dbj|BAB11155.1| contains similarity to unknown protein~gene_id:MOJ9.14 [Arabidopsis
thaliana]
Length = 1105
Score = 150 bits (380), Expect = 7e-36
Identities = 75/94 (79%), Positives = 86/94 (90%), Gaps = 1/94 (1%)
Frame = -2
Query: 548 DLEXLKEFFISGVDGLPRGVVENQVARVRHVIKLHGYETRELIEDLRSASGLEM-HGGKS 372
D+E LKEFFISG DGLPRGVVENQVARVR V+KLHGYETRELI+DLRS S LEM GGK
Sbjct: 1010 DVEVLKEFFISGGDGLPRGVVENQVARVRLVVKLHGYETRELIDDLRSRSSLEMQQGGKG 1069
Query: 371 KLGADSKTLLRILCHRSDSEASQFLKKQYQMPKS 270
KLGAD++TL+R+LCHR+DSEASQFLKKQY++P+S
Sbjct: 1070 KLGADTQTLVRVLCHRNDSEASQFLKKQYKIPRS 1103
>ref|NP_196314.2| putative protein; protein id: At5g06970.1, supported by cDNA:
gi_19699368 [Arabidopsis thaliana]
gi|19699369|gb|AAL91294.1| AT5g06970/MOJ9_14 [Arabidopsis
thaliana] gi|27363442|gb|AAO11640.1| At5g06970/MOJ9_14
[Arabidopsis thaliana]
Length = 1101
Score = 150 bits (380), Expect = 7e-36
Identities = 75/94 (79%), Positives = 86/94 (90%), Gaps = 1/94 (1%)
Frame = -2
Query: 548 DLEXLKEFFISGVDGLPRGVVENQVARVRHVIKLHGYETRELIEDLRSASGLEM-HGGKS 372
D+E LKEFFISG DGLPRGVVENQVARVR V+KLHGYETRELI+DLRS S LEM GGK
Sbjct: 1006 DVEVLKEFFISGGDGLPRGVVENQVARVRLVVKLHGYETRELIDDLRSRSSLEMQQGGKG 1065
Query: 371 KLGADSKTLLRILCHRSDSEASQFLKKQYQMPKS 270
KLGAD++TL+R+LCHR+DSEASQFLKKQY++P+S
Sbjct: 1066 KLGADTQTLVRVLCHRNDSEASQFLKKQYKIPRS 1099
>gb|AAK14418.1|AC087851_10 unknown protein [Oryza sativa]
Length = 1049
Score = 117 bits (292), Expect = 1e-25
Identities = 61/94 (64%), Positives = 72/94 (75%)
Frame = -2
Query: 548 DLEXLKEFFISGVDGLPRGVVENQVARVRHVIKLHGYETRELIEDLRSASGLEMHGGKSK 369
DLE LKEFFISG DGLPRG VEN V+RVR VI L ETR LI+DLR + G KSK
Sbjct: 957 DLEILKEFFISGGDGLPRGTVENLVSRVRPVIDLIKQETRVLIDDLREVT----QGAKSK 1012
Query: 368 LGADSKTLLRILCHRSDSEASQFLKKQYQMPKSS 267
G DSKTLLR+LCHR+DSEAS ++KKQ+++P S+
Sbjct: 1013 FGTDSKTLLRVLCHRNDSEASHYVKKQFKIPSSA 1046
>ref|NP_192904.1| hypothetical protein; protein id: At4g11670.1 [Arabidopsis thaliana]
gi|7452474|pir||T04214 hypothetical protein T5C23.100 -
Arabidopsis thaliana gi|4539458|emb|CAB39938.1|
hypothetical protein [Arabidopsis thaliana]
gi|7267867|emb|CAB78210.1| hypothetical protein
[Arabidopsis thaliana]
Length = 998
Score = 80.1 bits (196), Expect = 2e-14
Identities = 41/96 (42%), Positives = 64/96 (65%), Gaps = 2/96 (2%)
Frame = -2
Query: 548 DLEXLKEFFISGVDGLPRGVVENQVARVRHVIKLHGYETRELIEDLRSASGLEMHGGKSK 369
DL LKEFFI+ +GLPR +VE + + + ++ L+ E+ LI+ L +AS L G S+
Sbjct: 852 DLSILKEFFIADGEGLPRSLVEQEAKQAKEILDLYSLESDMLIQMLMTASELINMGVSSE 911
Query: 368 LGA--DSKTLLRILCHRSDSEASQFLKKQYQMPKSS 267
D++TL+R+LCH+ D AS+FLK+QY++P S+
Sbjct: 912 QRRLEDAQTLVRVLCHKKDRNASKFLKRQYELPMST 947
>ref|NP_180900.1| unknown protein; protein id: At2g33420.1 [Arabidopsis thaliana]
gi|25408269|pir||C84745 hypothetical protein At2g33420
[imported] - Arabidopsis thaliana
gi|2459424|gb|AAB80659.1| unknown protein [Arabidopsis
thaliana]
Length = 1039
Score = 66.2 bits (160), Expect = 2e-10
Identities = 42/107 (39%), Positives = 57/107 (53%), Gaps = 15/107 (14%)
Frame = -2
Query: 548 DLEXLKEFFISGVDGL-PRGVVENQVARVRHVIKLHGYETRELIEDLR----SASGLEMH 384
D E LK F + +GL P VV+ + V VI+L T +L+ED SG+ M
Sbjct: 931 DFENLKRVFCTCGEGLIPEEVVDREAETVEGVIQLMSQPTEQLMEDFSIVTCETSGMGMV 990
Query: 383 GGKSKLG----------ADSKTLLRILCHRSDSEASQFLKKQYQMPK 273
G KL +D T+LR+LCHR+D A+QFLKK +Q+PK
Sbjct: 991 GSGQKLPMPPTTGRWNRSDPNTILRVLCHRNDRVANQFLKKSFQLPK 1037
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 432,402,702
Number of Sequences: 1393205
Number of extensions: 8527496
Number of successful extensions: 32198
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 31531
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32177
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)