Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000646A_C01 KMC000646A_c01
(591 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
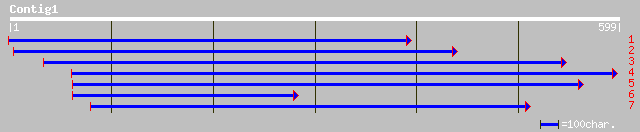
gb|AAK70633.1|AC091238_11 Putative gag-pol polyprotein [Oryza sa... 82 2e-33
pir||T17429 gag-pol polyprotein - maize copia-like retrotranspos... 70 2e-22
gb|AAD12994.1| gag-pol polyprotein [Zea mays] 70 2e-22
dbj|BAB89089.1| putative gag-pol polyprotein [Oryza sativa (japo... 79 3e-14
emb|CAD39757.1| OSJNBa0059D20.12 [Oryza sativa (japonica cultiva... 76 3e-13
>gb|AAK70633.1|AC091238_11 Putative gag-pol polyprotein [Oryza sativa]
gi|21321753|gb|AAM47288.1|AC122146_7 Putative gag-pol
polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1225
Score = 82.4 bits (202), Expect(2) = 2e-33
Identities = 42/117 (35%), Positives = 59/117 (49%), Gaps = 20/117 (17%)
Frame = -2
Query: 590 FKISYNCPKEKWSLNELISFCVQEEERLKQERKESAHFVSTS------------------ 465
F I+YN K KW ++ELI+ CVQEEERLK ER + A+ S
Sbjct: 185 FTINYNAMKIKWGIDELIAMCVQEEERLKAERVDHANLFKHSEKKKYKKFKKEYLKPKPI 244
Query: 464 --KDKGKRKKTVEPKNEAADAPAQKKQKEDDTCYFCNVSGHMKKKCTKYHAWRARKG 300
K+KG+ + + ++K+KE D CYFC SGH +K C + W ++KG
Sbjct: 245 QFKEKGQSSSQSQQNKSGGNPYTEEKEKEKDGCYFCGKSGHRQKDCIGFMRWLSKKG 301
Score = 81.6 bits (200), Expect(2) = 2e-33
Identities = 46/101 (45%), Positives = 59/101 (57%)
Frame = -3
Query: 304 KGCLSYRKLNDVERYIYVGDGKTVEVEAIGHFRLLLCTGFYLDLKDTFVVPSFRRNLISV 125
+G R L ER I V +G EVEAIG F L L TGF L L D VPS +RNL+SV
Sbjct: 336 QGFAMRRTLRRGERRIRVANGVETEVEAIGDFTLTLHTGFKLQLHDVLYVPSMKRNLVSV 395
Query: 124 SNLDKSGYSCSFGNSKAELSFNSNIVGTGSLIGYDNLYLLS 2
S LD GY C+FGN++ + ++ VG +G + LY +S
Sbjct: 396 SRLDNGGYYCTFGNNRCIIMHDNKEVGLA--VGREQLYQIS 434
>pir||T17429 gag-pol polyprotein - maize copia-like retrotransposon Sto-4
gi|4234852|gb|AAD12997.1| gag-pol polyprotein [Zea mays]
Length = 1406
Score = 70.1 bits (170), Expect(2) = 2e-22
Identities = 44/101 (43%), Positives = 55/101 (53%)
Frame = -3
Query: 304 KGCLSYRKLNDVERYIYVGDGKTVEVEAIGHFRLLLCTGFYLDLKDTFVVPSFRRNLISV 125
+G + ++L R I V +G VEAIG L L GF L L+D VPS RRNLISV
Sbjct: 316 QGLRTSQRLPKGRRTIRVANGVEAAVEAIGDIHLKLVNGFVLLLRDVLYVPSLRRNLISV 375
Query: 124 SNLDKSGYSCSFGNSKAELSFNSNIVGTGSLIGYDNLYLLS 2
S LD C FG+ + + F++ VG I D LYLLS
Sbjct: 376 SRLDDQHIHCHFGDRQCVIQFDNKDVGLA--IRRDMLYLLS 414
Score = 57.4 bits (137), Expect(2) = 2e-22
Identities = 34/110 (30%), Positives = 57/110 (50%), Gaps = 13/110 (11%)
Frame = -2
Query: 590 FKISYNCP-KEKWSLNELISFCVQEEERLKQERKESAHFV------STSKDKGKRKKTVE 432
F I YN +KW++++L++ CVQEEERLKQ +S + + K K KK +
Sbjct: 173 FHIQYNTSVTDKWNIDQLLAQCVQEEERLKQ-TGDSVNLIKGNVPRQNKNSKKKFKKQGK 231
Query: 431 PKNEAADAPAQKKQK------EDDTCYFCNVSGHMKKKCTKYHAWRARKG 300
N+A+ + + ++ DTC +C +GH K+KC ++ + G
Sbjct: 232 QNNQASSSNNNQPRQGGGFSVPPDTCLYCKKTGHYKRKCPEFLQYLLESG 281
>gb|AAD12994.1| gag-pol polyprotein [Zea mays]
Length = 595
Score = 70.1 bits (170), Expect(2) = 2e-22
Identities = 44/101 (43%), Positives = 55/101 (53%)
Frame = -3
Query: 304 KGCLSYRKLNDVERYIYVGDGKTVEVEAIGHFRLLLCTGFYLDLKDTFVVPSFRRNLISV 125
+G + ++L R I V +G VEAIG L L GF L L+D VPS RRNLISV
Sbjct: 316 QGLRTSQRLPKGRRTIRVANGVEAAVEAIGDIHLKLVNGFVLLLRDVLYVPSLRRNLISV 375
Query: 124 SNLDKSGYSCSFGNSKAELSFNSNIVGTGSLIGYDNLYLLS 2
S LD C FG+ + + F++ VG I D LYLLS
Sbjct: 376 SRLDDQHIHCHFGDRQCVIQFDNKDVGLA--IRRDMLYLLS 414
Score = 57.4 bits (137), Expect(2) = 2e-22
Identities = 34/110 (30%), Positives = 57/110 (50%), Gaps = 13/110 (11%)
Frame = -2
Query: 590 FKISYNCP-KEKWSLNELISFCVQEEERLKQERKESAHFV------STSKDKGKRKKTVE 432
F I YN +KW++++L++ CVQEEERLKQ +S + + K K KK +
Sbjct: 173 FHIQYNTSVTDKWNIDQLLAQCVQEEERLKQ-TGDSVNLIKGNVPRQNKNSKKKFKKQGK 231
Query: 431 PKNEAADAPAQKKQK------EDDTCYFCNVSGHMKKKCTKYHAWRARKG 300
N+A+ + + ++ DTC +C +GH K+KC ++ + G
Sbjct: 232 QNNQASSSNNNQPRQGGSFSVPPDTCLYCKKTGHYKRKCPEFLQYLLESG 281
>dbj|BAB89089.1| putative gag-pol polyprotein [Oryza sativa (japonica
cultivar-group)] gi|21328072|dbj|BAC00656.1| putative
gag-pol polyprotein [Oryza sativa (japonica
cultivar-group)]
Length = 1397
Score = 79.3 bits (194), Expect = 3e-14
Identities = 49/101 (48%), Positives = 56/101 (54%)
Frame = -3
Query: 304 KGCLSYRKLNDVERYIYVGDGKTVEVEAIGHFRLLLCTGFYLDLKDTFVVPSFRRNLISV 125
K S R E I V +G +VEA+G L L GF L L+D F VPS +RNLISV
Sbjct: 324 KAFRSTRTTQRRESTIRVANGVEEKVEAVGDLPLELSNGFILLLRDVFYVPSLQRNLISV 383
Query: 124 SNLDKSGYSCSFGNSKAELSFNSNIVGTGSLIGYDNLYLLS 2
S LD GY C FGN K EL N+ +G L D LYLLS
Sbjct: 384 SKLDFDGYDCRFGNGKCELWHNNACIGLAVL--RDELYLLS 422
Score = 65.9 bits (159), Expect = 4e-10
Identities = 35/91 (38%), Positives = 51/91 (55%), Gaps = 2/91 (2%)
Frame = -2
Query: 590 FKISYNCPKEKWSLNELISFCVQEEERLKQERKESAHFVSTSKDKGKRKKTVEPKNEAAD 411
F ++YN EKW+ +LI+ CVQEEER+K+ S ++V KDK K K+ K + +
Sbjct: 193 FIVNYNISPEKWNFEKLIANCVQEEERIKESNGGSINYV---KDKKKNHKSPTSKGKQSQ 249
Query: 410 APAQKKQ--KEDDTCYFCNVSGHMKKKCTKY 324
Q++Q E D C C +GH KK C +
Sbjct: 250 HLPQQQQFAVEKDQCLHCKKTGHYKKDCPDF 280
>emb|CAD39757.1| OSJNBa0059D20.12 [Oryza sativa (japonica cultivar-group)]
Length = 497
Score = 76.3 bits (186), Expect = 3e-13
Identities = 47/101 (46%), Positives = 57/101 (55%)
Frame = -3
Query: 304 KGCLSYRKLNDVERYIYVGDGKTVEVEAIGHFRLLLCTGFYLDLKDTFVVPSFRRNLISV 125
K S R E I V +G +VEA G L L GF L L+D F VPSF+RNLISV
Sbjct: 132 KAFCSMRTTQRSESSIGVANGVEEKVEAAGDLCLELTNGFTLLLRDVFYVPSFQRNLISV 191
Query: 124 SNLDKSGYSCSFGNSKAELSFNSNIVGTGSLIGYDNLYLLS 2
S LD GY C FG+ K EL + + +G ++ D LYLLS
Sbjct: 192 SKLDFDGYDCRFGSGKCELWYYNACIGL--VVLQDELYLLS 230
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 504,644,052
Number of Sequences: 1393205
Number of extensions: 11082871
Number of successful extensions: 44584
Number of sequences better than 10.0: 330
Number of HSP's better than 10.0 without gapping: 39783
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44081
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)