Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000635A_C01 KMC000635A_c01
(614 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
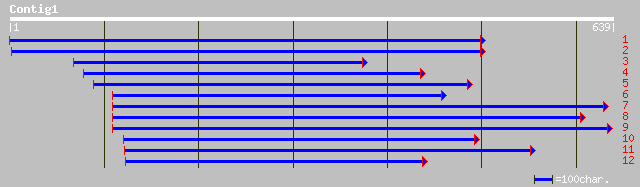
gb|AAD41796.1| precursor monofunctional aspartokinase [Glycine max] 134 7e-31
gb|AAB63104.1| lysine-sensitive aspartate kinase [Arabidopsis th... 109 3e-23
ref|NP_196910.1| lysine-sensitive aspartate kinase (gb|AAB63104.... 109 3e-23
emb|CAC06395.1| aspartate kinase [Arabidopsis thaliana] 109 3e-23
ref|NP_186851.1| putative aspartate kinase; protein id: At3g0202... 108 4e-23
>gb|AAD41796.1| precursor monofunctional aspartokinase [Glycine max]
Length = 564
Score = 134 bits (338), Expect = 7e-31
Identities = 75/85 (88%), Positives = 78/85 (91%), Gaps = 4/85 (4%)
Frame = -1
Query: 614 SIISLIGNVQRSSLILEKAFRVLRSLGVTVQMISQGASKVNISLVVNDNEAEQCVRALHS 435
SIISLIGNVQRSSLILE+ RVLR+LGVTVQMISQGASKVNISLVVND+EAEQCVRALHS
Sbjct: 480 SIISLIGNVQRSSLILERLSRVLRTLGVTVQMISQGASKVNISLVVNDSEAEQCVRALHS 539
Query: 434 AFFESELSELE----NGNGSVAELS 372
AFFESELSELE NGNGSV ELS
Sbjct: 540 AFFESELSELEMDYKNGNGSVDELS 564
>gb|AAB63104.1| lysine-sensitive aspartate kinase [Arabidopsis thaliana]
Length = 544
Score = 109 bits (272), Expect = 3e-23
Identities = 56/66 (84%), Positives = 61/66 (91%)
Frame = -1
Query: 614 SIISLIGNVQRSSLILEKAFRVLRSLGVTVQMISQGASKVNISLVVNDNEAEQCVRALHS 435
SIISLIGNVQ+SSLILEK F+V RS GV VQMISQGASKVNISL+VND EAEQCVRALHS
Sbjct: 478 SIISLIGNVQKSSLILEKVFQVFRSNGVNVQMISQGASKVNISLIVNDEEAEQCVRALHS 537
Query: 434 AFFESE 417
AFFE++
Sbjct: 538 AFFETD 543
>ref|NP_196910.1| lysine-sensitive aspartate kinase (gb|AAB63104.1); protein id:
At5g14060.1, supported by cDNA: gi_2257742 [Arabidopsis
thaliana] gi|9757787|dbj|BAB08285.1| lysine-sensitive
aspartate kinase [Arabidopsis thaliana]
Length = 544
Score = 109 bits (272), Expect = 3e-23
Identities = 56/66 (84%), Positives = 61/66 (91%)
Frame = -1
Query: 614 SIISLIGNVQRSSLILEKAFRVLRSLGVTVQMISQGASKVNISLVVNDNEAEQCVRALHS 435
SIISLIGNVQ+SSLILEK F+V RS GV VQMISQGASKVNISL+VND EAEQCVRALHS
Sbjct: 478 SIISLIGNVQKSSLILEKVFQVFRSNGVNVQMISQGASKVNISLIVNDEEAEQCVRALHS 537
Query: 434 AFFESE 417
AFFE++
Sbjct: 538 AFFETD 543
>emb|CAC06395.1| aspartate kinase [Arabidopsis thaliana]
Length = 544
Score = 109 bits (272), Expect = 3e-23
Identities = 56/66 (84%), Positives = 61/66 (91%)
Frame = -1
Query: 614 SIISLIGNVQRSSLILEKAFRVLRSLGVTVQMISQGASKVNISLVVNDNEAEQCVRALHS 435
SIISLIGNVQ+SSLILEK F+V RS GV VQMISQGASKVNISL+VND EAEQCVRALHS
Sbjct: 478 SIISLIGNVQKSSLILEKVFQVFRSNGVNVQMISQGASKVNISLIVNDEEAEQCVRALHS 537
Query: 434 AFFESE 417
AFFE++
Sbjct: 538 AFFETD 543
>ref|NP_186851.1| putative aspartate kinase; protein id: At3g02020.1, supported by
cDNA: 6203. [Arabidopsis thaliana]
gi|6091740|gb|AAF03452.1|AC010797_28 putative aspartate
kinase [Arabidopsis thaliana]
gi|6513929|gb|AAF14833.1|AC011664_15 putative aspartate
kinase [Arabidopsis thaliana] gi|21593967|gb|AAM65905.1|
putative aspartate kinase [Arabidopsis thaliana]
Length = 559
Score = 108 bits (271), Expect = 4e-23
Identities = 55/69 (79%), Positives = 62/69 (89%)
Frame = -1
Query: 614 SIISLIGNVQRSSLILEKAFRVLRSLGVTVQMISQGASKVNISLVVNDNEAEQCVRALHS 435
SIISLIGNVQRSS ILEK FRVLR+ G+ VQMISQGASKVNISL+VND+EAE CV+ALHS
Sbjct: 477 SIISLIGNVQRSSFILEKGFRVLRTNGINVQMISQGASKVNISLIVNDDEAEHCVKALHS 536
Query: 434 AFFESELSE 408
AFFE++ E
Sbjct: 537 AFFETDTCE 545
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,930,127
Number of Sequences: 1393205
Number of extensions: 9665312
Number of successful extensions: 18920
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 18469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18910
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24854530794
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)