Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000626A_C01 KMC000626A_c01
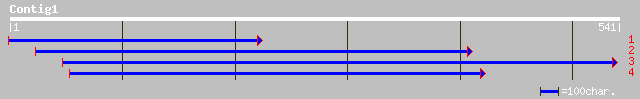
(540 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||H86327 protein F18O14.25 [imported] - Arabidopsis thaliana ... 74 9e-13
ref|NP_564086.1| hypothetical protein; protein id: At1g19485.1 [... 74 9e-13
dbj|BAB63709.1| P0683F02.12 [Oryza sativa (japonica cultivar-gro... 69 4e-11
dbj|BAC65402.1| OSJNBa0075N02.16 [Oryza sativa (japonica cultiva... 65 5e-10
gb|AAC09349.1| unknown [Homo sapiens] 33 2.9
>pir||H86327 protein F18O14.25 [imported] - Arabidopsis thaliana
gi|8778423|gb|AAF79431.1|AC025808_13 F18O14.25
[Arabidopsis thaliana]
Length = 1314
Score = 74.3 bits (181), Expect = 9e-13
Identities = 30/41 (73%), Positives = 36/41 (87%)
Frame = -3
Query: 388 GVYLKSFLPKMVALHRVRWNMNMGSERWLCYGGASGVLRCQ 266
G+ + F PKMVA+HRVRWNMN GSERWLCYGGA+G++RCQ
Sbjct: 1262 GMKAEGFPPKMVAMHRVRWNMNKGSERWLCYGGAAGIVRCQ 1302
>ref|NP_564086.1| hypothetical protein; protein id: At1g19485.1 [Arabidopsis
thaliana]
Length = 815
Score = 74.3 bits (181), Expect = 9e-13
Identities = 30/41 (73%), Positives = 36/41 (87%)
Frame = -3
Query: 388 GVYLKSFLPKMVALHRVRWNMNMGSERWLCYGGASGVLRCQ 266
G+ + F PKMVA+HRVRWNMN GSERWLCYGGA+G++RCQ
Sbjct: 763 GMKAEGFPPKMVAMHRVRWNMNKGSERWLCYGGAAGIVRCQ 803
>dbj|BAB63709.1| P0683F02.12 [Oryza sativa (japonica cultivar-group)]
Length = 1132
Score = 68.9 bits (167), Expect = 4e-11
Identities = 27/35 (77%), Positives = 32/35 (91%)
Frame = -3
Query: 370 FLPKMVALHRVRWNMNMGSERWLCYGGASGVLRCQ 266
F PK VALHR+RWNMN GSE+WLCYGGA+G++RCQ
Sbjct: 1096 FPPKAVALHRLRWNMNKGSEKWLCYGGAAGIIRCQ 1130
>dbj|BAC65402.1| OSJNBa0075N02.16 [Oryza sativa (japonica cultivar-group)]
Length = 1193
Score = 65.1 bits (157), Expect = 5e-10
Identities = 28/45 (62%), Positives = 36/45 (79%), Gaps = 1/45 (2%)
Frame = -3
Query: 370 FLPKMVALHRVRWNMNMGSERWLCYGGASGVLRC-QHD*VISYRI 239
F PK VALHR+RWNMN GSE+WLCYGGA+G++R H ++ YR+
Sbjct: 1115 FPPKAVALHRLRWNMNKGSEKWLCYGGAAGIIRVYPHTALVWYRL 1159
>gb|AAC09349.1| unknown [Homo sapiens]
Length = 292
Score = 32.7 bits (73), Expect = 2.9
Identities = 13/30 (43%), Positives = 22/30 (73%)
Frame = -3
Query: 361 KMVALHRVRWNMNMGSERWLCYGGASGVLR 272
++ A+H+VR++ N+ S WL GG SG++R
Sbjct: 214 QLEAIHKVRFSPNLDSYGWLVSGGQSGLVR 243
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 435,110,873
Number of Sequences: 1393205
Number of extensions: 9211285
Number of successful extensions: 18702
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 18270
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18687
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)