Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000611A_C01 KMC000611A_c01
(542 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
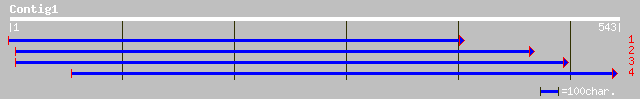
pir||T05692 hypothetical protein F20M13.200 - Arabidopsis thalia... 191 5e-48
ref|NP_568045.1| putative protein; protein id: At4g38640.1 [Arab... 191 5e-48
dbj|BAB62997.1| hypothetical protein [Macaca fascicularis] 40 0.024
dbj|BAB63011.1| hypothetical protein [Macaca fascicularis] 39 0.031
dbj|BAC04084.1| unnamed protein product [Homo sapiens] 37 0.20
>pir||T05692 hypothetical protein F20M13.200 - Arabidopsis thaliana
gi|4467151|emb|CAB37520.1| putative protein [Arabidopsis
thaliana] gi|7270847|emb|CAB80528.1| putative protein
[Arabidopsis thaliana]
Length = 523
Score = 191 bits (485), Expect = 5e-48
Identities = 91/126 (72%), Positives = 107/126 (84%)
Frame = -3
Query: 540 EAYCSSXRMTYELLRRNLLSAAFVETISSRLLAGIVFVLSAIYTIVVCVILKYGTNLGSD 361
EAYC+S +MTYELLRRNLLSA FVET+S+R+L GIVFVLSA Y + +L+ +NLG D
Sbjct: 392 EAYCTSAKMTYELLRRNLLSAVFVETVSTRILTGIVFVLSAAYAVATWAVLRGVSNLGID 451
Query: 360 SYFVAAMAWVLLIIVLGFLVHVLDDVIDTVYVCYAIDRDRGEVCKQDVHEVYVHLPISRS 181
SY VA +AWVLLI++L F VHVLDDVIDT+YVCYAIDRD+G+VCKQ+VHEVYVHLPISRS
Sbjct: 452 SYVVAVLAWVLLIVILAFFVHVLDDVIDTIYVCYAIDRDKGDVCKQEVHEVYVHLPISRS 511
Query: 180 LRQSNI 163
R S I
Sbjct: 512 TRSSLI 517
>ref|NP_568045.1| putative protein; protein id: At4g38640.1 [Arabidopsis thaliana]
gi|16649077|gb|AAL24390.1| putative protein [Arabidopsis
thaliana] gi|21387167|gb|AAM47987.1| putative protein
[Arabidopsis thaliana]
Length = 556
Score = 191 bits (485), Expect = 5e-48
Identities = 91/126 (72%), Positives = 107/126 (84%)
Frame = -3
Query: 540 EAYCSSXRMTYELLRRNLLSAAFVETISSRLLAGIVFVLSAIYTIVVCVILKYGTNLGSD 361
EAYC+S +MTYELLRRNLLSA FVET+S+R+L GIVFVLSA Y + +L+ +NLG D
Sbjct: 425 EAYCTSAKMTYELLRRNLLSAVFVETVSTRILTGIVFVLSAAYAVATWAVLRGVSNLGID 484
Query: 360 SYFVAAMAWVLLIIVLGFLVHVLDDVIDTVYVCYAIDRDRGEVCKQDVHEVYVHLPISRS 181
SY VA +AWVLLI++L F VHVLDDVIDT+YVCYAIDRD+G+VCKQ+VHEVYVHLPISRS
Sbjct: 485 SYVVAVLAWVLLIVILAFFVHVLDDVIDTIYVCYAIDRDKGDVCKQEVHEVYVHLPISRS 544
Query: 180 LRQSNI 163
R S I
Sbjct: 545 TRSSLI 550
>dbj|BAB62997.1| hypothetical protein [Macaca fascicularis]
Length = 468
Score = 39.7 bits (91), Expect = 0.024
Identities = 33/137 (24%), Positives = 64/137 (46%), Gaps = 10/137 (7%)
Frame = -3
Query: 537 AYCSSXRMTYELLRRNLLSAA-------FVETISSRLLAGIVFVLSAIYTIVVCVILKYG 379
++C S + + LL RN+L A FV + L+AG + VL+ ++ ++ G
Sbjct: 334 SFCRSAKDAFNLLMRNVLKVAVTDEVTYFVLFLGKILVAGSIGVLAFLFFTQRLPVIAQG 393
Query: 378 TNLGSDSYFVAAMAWVLL---IIVLGFLVHVLDDVIDTVYVCYAIDRDRGEVCKQDVHEV 208
S +Y+ + V+L +I GF V ++T+++C+ D +R +
Sbjct: 394 P--ASLNYYWVPLLTVILGSYLIAHGFF-SVYAMCVETIFICFLEDLERND--GSTARPY 448
Query: 207 YVHLPISRSLRQSNIIT 157
YV P+ + ++ N+ T
Sbjct: 449 YVSQPLLKIFQEENLQT 465
>dbj|BAB63011.1| hypothetical protein [Macaca fascicularis]
Length = 717
Score = 39.3 bits (90), Expect = 0.031
Identities = 33/136 (24%), Positives = 63/136 (46%), Gaps = 10/136 (7%)
Frame = -3
Query: 534 YCSSXRMTYELLRRNLLSAA-------FVETISSRLLAGIVFVLSAIYTIVVCVILKYGT 376
+C S + + LL RN+L A FV + L+AG + VL+ ++ ++ G
Sbjct: 584 FCRSAKDAFNLLMRNVLKVAVTDEVTYFVLFLGKILVAGSIGVLAFLFFTQRLPVIAQGP 643
Query: 375 NLGSDSYFVAAMAWVLL---IIVLGFLVHVLDDVIDTVYVCYAIDRDRGEVCKQDVHEVY 205
S +Y+ + V+L +I GF V ++T+++C+ D +R + Y
Sbjct: 644 --ASLNYYWVPLLTVILGSYLIAHGFF-SVYAMCVETIFICFLEDLERND--GSTARPYY 698
Query: 204 VHLPISRSLRQSNIIT 157
V P+ + ++ N+ T
Sbjct: 699 VSQPLLKIFQEENLQT 714
>dbj|BAC04084.1| unnamed protein product [Homo sapiens]
Length = 717
Score = 36.6 bits (83), Expect = 0.20
Identities = 31/133 (23%), Positives = 60/133 (44%), Gaps = 10/133 (7%)
Frame = -3
Query: 534 YCSSXRMTYELLRRNLLSAA-------FVETISSRLLAGIVFVLSAIYTIVVCVILKYGT 376
+C S + + LL RN+L A FV + L+AG + VL+ ++ ++ G
Sbjct: 584 FCRSAKDAFNLLMRNVLKVAVTDEVTYFVLFLGKLLVAGSIGVLAFLFFTQRLPVIAQGP 643
Query: 375 NLGSDSYFVAAMAWVLL---IIVLGFLVHVLDDVIDTVYVCYAIDRDRGEVCKQDVHEVY 205
S +Y+ + V+ +I GF V ++T+++C+ D +R + Y
Sbjct: 644 --ASLNYYWVPLLTVIFGSYLIAHGFF-SVYAMCVETIFICFLEDLERND--GSTARPYY 698
Query: 204 VHLPISRSLRQSN 166
V P+ + ++ N
Sbjct: 699 VSQPLLKIFQEEN 711
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 462,184,380
Number of Sequences: 1393205
Number of extensions: 10151063
Number of successful extensions: 25237
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 24465
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25216
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18750593680
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)