Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000605A_C01 KMC000605A_c01
(541 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
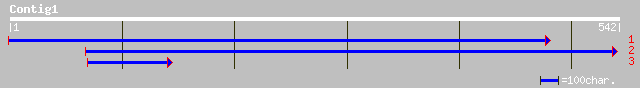
gb|AAN05494.1| Putative F-box protein [Oryza sativa (japonica cu... 103 2e-21
ref|NP_565400.1| F-box protein family, AtFBL10; protein id: At2g... 94 1e-18
gb|AAM61680.1| unknown [Arabidopsis thaliana] 94 1e-18
gb|AAG25917.1|AF190427_1 putative GAL4-like transcriptional acti... 44 0.001
gb|EAA12317.1| agCP11454 [Anopheles gambiae str. PEST] 42 0.006
>gb|AAN05494.1| Putative F-box protein [Oryza sativa (japonica cultivar-group)]
Length = 622
Score = 103 bits (256), Expect = 2e-21
Identities = 47/78 (60%), Positives = 58/78 (74%)
Frame = -2
Query: 540 YFPRLKWLGVTGSVNRDMVDALARSRPFLHVACHGEELGADPYDISDGLYTHDYDEVDEF 361
YFPRL+WLGVTGS+NR MVDAL RSRPFLH+AC GEELG +D S Y HD D++DE
Sbjct: 544 YFPRLRWLGVTGSLNRVMVDALVRSRPFLHMACRGEELGTFNWDRSSDWYRHDDDDLDEL 603
Query: 360 EQWLLEADIDTDDEEMVD 307
EQW+L + +D E + +
Sbjct: 604 EQWILNGEPVSDTETITE 621
>ref|NP_565400.1| F-box protein family, AtFBL10; protein id: At2g17020.1, supported
by cDNA: gi_13605808 [Arabidopsis thaliana]
gi|25372906|pir||B84547 hypothetical protein At2g17020
[imported] - Arabidopsis thaliana
gi|13605809|gb|AAK32890.1|AF367303_1 At2g17020
[Arabidopsis thaliana] gi|22137200|gb|AAM91445.1|
At2g17020/At2g17020 [Arabidopsis thaliana]
Length = 656
Score = 93.6 bits (231), Expect = 1e-18
Identities = 50/86 (58%), Positives = 57/86 (66%), Gaps = 2/86 (2%)
Frame = -2
Query: 540 YFPRLKWLGVTGSVNRDMVDALARSRPFLHVACHGEELGADPYDISDGLYTHDYDEV--D 367
+FPRLKWLG+TGSVNRD+VDALAR RP L V+C GEELG D D D H + E D
Sbjct: 567 FFPRLKWLGITGSVNRDIVDALARRRPHLQVSCRGEELGNDGEDDWDSADIHQHIEAQED 626
Query: 366 EFEQWLLEADIDTDDEEMVDAENNEE 289
E EQW+L D D EM DAE+ E
Sbjct: 627 ELEQWILG---DEGDVEMEDAEDESE 649
>gb|AAM61680.1| unknown [Arabidopsis thaliana]
Length = 130
Score = 93.6 bits (231), Expect = 1e-18
Identities = 50/86 (58%), Positives = 57/86 (66%), Gaps = 2/86 (2%)
Frame = -2
Query: 540 YFPRLKWLGVTGSVNRDMVDALARSRPFLHVACHGEELGADPYDISDGLYTHDYDEV--D 367
+FPRLKWLG+TGSVNRD+VDALAR RP L V+C GEELG D D D H + E D
Sbjct: 41 FFPRLKWLGITGSVNRDIVDALARRRPHLQVSCRGEELGNDGEDDWDSADIHQHIEAQED 100
Query: 366 EFEQWLLEADIDTDDEEMVDAENNEE 289
E EQW+L D D EM DAE+ E
Sbjct: 101 ELEQWILG---DEGDVEMEDAEDESE 123
>gb|AAG25917.1|AF190427_1 putative GAL4-like transcriptional activator [Colletotrichum
lindemuthianum]
Length = 746
Score = 43.9 bits (102), Expect = 0.001
Identities = 25/57 (43%), Positives = 28/57 (48%), Gaps = 7/57 (12%)
Frame = +3
Query: 303 QHQPFLHRQYQYQLQAATAQIH------PPHHNHVYISHLRYHTDQPRAPH-HGMPH 452
QHQP H+Q Q+Q Q Q PP H H + SH Y T QP PH H PH
Sbjct: 629 QHQPQQHQQQQHQQQQHQLQHQQFSPGPPPPHPHHHQSHASYPTPQPPQPHPHHQPH 685
>gb|EAA12317.1| agCP11454 [Anopheles gambiae str. PEST]
Length = 268
Score = 41.6 bits (96), Expect = 0.006
Identities = 21/50 (42%), Positives = 26/50 (52%)
Frame = +3
Query: 303 QHQPFLHRQYQYQLQAATAQIHPPHHNHVYISHLRYHTDQPRAPHHGMPH 452
QH H Q+Q Q Q +A PPHH+H + H +H P HGMPH
Sbjct: 70 QHSQMNHHQHQQQQQPPSAP--PPHHHHHH--HHAHHPAASAGPLHGMPH 115
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 455,659,330
Number of Sequences: 1393205
Number of extensions: 9526834
Number of successful extensions: 56299
Number of sequences better than 10.0: 389
Number of HSP's better than 10.0 without gapping: 32521
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43991
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)