Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000603A_C01 KMC000603A_c01
(1000 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
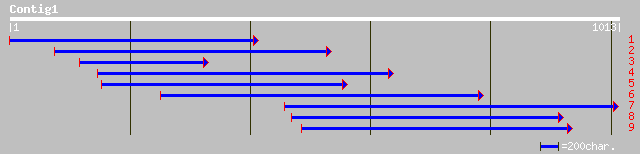
ref|NP_181543.1| putative CCCH-type zinc finger protein; protein... 127 2e-28
ref|NP_567030.1| putative protein; protein id: At3g55980.1, supp... 115 1e-24
pir||T49210 hypothetical protein F27K19.160 - Arabidopsis thalia... 115 1e-24
ref|NP_200670.1| putative protein; protein id: At5g58620.1, supp... 113 5e-24
gb|AAO38462.1| unknown protein [Oryza sativa (japonica cultivar-... 73 8e-12
>ref|NP_181543.1| putative CCCH-type zinc finger protein; protein id: At2g40140.1,
supported by cDNA: gi_20260233 [Arabidopsis thaliana]
gi|25370571|pir||G84825 probable CCCH-type zinc finger
protein [imported] - Arabidopsis thaliana
gi|4587989|gb|AAD25930.1|AF085279_3 hypothetical
Cys-3-His zinc finger protein [Arabidopsis thaliana]
gi|20260234|gb|AAM13015.1| putative CCCH-type zinc
finger protein [Arabidopsis thaliana]
gi|22136518|gb|AAM91337.1| putative CCCH-type zinc
finger protein [Arabidopsis thaliana]
Length = 597
Score = 127 bits (320), Expect = 2e-28
Identities = 75/169 (44%), Positives = 95/169 (55%), Gaps = 18/169 (10%)
Frame = -3
Query: 992 SNIPSSPVRKPSSLGFDSSSAVAAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNP 813
++ PSSPVR+P GF+SS+A+AAAVMN+RS+AFAKRS SF
Sbjct: 446 NHYPSSPVRQPPPHGFESSAAMAAAVMNARSSAFAKRSLSF------------------K 487
Query: 812 SCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDV 633
V+S SDW SP+GKL+WG+ DEL+KLR+SASFG N P A EPDV
Sbjct: 488 PAPVASNVSDWGSPNGKLEWGMQRDELNKLRRSASFGIHGNNNNSVSRP-ARDYSDEPDV 546
Query: 632 SWVHSLVKDDL------------------SKEMLPPWVEQLYIEQEQMV 540
SWV+SLVK++ K LP W EQ+YI+ EQ +
Sbjct: 547 SWVNSLVKENAPERVNERVGNTVNGAASRDKFKLPSWAEQMYIDHEQQI 595
>ref|NP_567030.1| putative protein; protein id: At3g55980.1, supported by cDNA:
gi_15810486 [Arabidopsis thaliana]
gi|15810487|gb|AAL07131.1| unknown protein [Arabidopsis
thaliana]
Length = 580
Score = 115 bits (287), Expect = 1e-24
Identities = 70/159 (44%), Positives = 87/159 (54%), Gaps = 11/159 (6%)
Frame = -3
Query: 983 PSSPVRKPSSL-GFDSSSAVAAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSC 807
PSSPVR+P S GF+SS+A A AVM +RS AFAKRS SF A
Sbjct: 440 PSSPVRQPPSQHGFESSAAAAVAVMKARSTAFAKRSLSFKPATQAAPQ------------ 487
Query: 806 RVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSW 627
S SDW SP+GKL+WG+ G+EL+K+R+S SFG N + A EPDVSW
Sbjct: 488 ---SNLSDWGSPNGKLEWGMKGEELNKMRRSVSFGIHGN----NNNNAARDYRDEPDVSW 540
Query: 626 VHSLVKDDL----------SKEMLPPWVEQLYIEQEQMV 540
V+SLVKD + + W EQ+Y E+EQ V
Sbjct: 541 VNSLVKDSTVVSERSFGMNERVRIMSWAEQMYREKEQTV 579
>pir||T49210 hypothetical protein F27K19.160 - Arabidopsis thaliana
gi|7573493|emb|CAB87852.1| putative protein [Arabidopsis
thaliana]
Length = 586
Score = 115 bits (287), Expect = 1e-24
Identities = 70/159 (44%), Positives = 87/159 (54%), Gaps = 11/159 (6%)
Frame = -3
Query: 983 PSSPVRKPSSL-GFDSSSAVAAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSC 807
PSSPVR+P S GF+SS+A A AVM +RS AFAKRS SF A
Sbjct: 446 PSSPVRQPPSQHGFESSAAAAVAVMKARSTAFAKRSLSFKPATQAAPQ------------ 493
Query: 806 RVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSW 627
S SDW SP+GKL+WG+ G+EL+K+R+S SFG N + A EPDVSW
Sbjct: 494 ---SNLSDWGSPNGKLEWGMKGEELNKMRRSVSFGIHGN----NNNNAARDYRDEPDVSW 546
Query: 626 VHSLVKDDL----------SKEMLPPWVEQLYIEQEQMV 540
V+SLVKD + + W EQ+Y E+EQ V
Sbjct: 547 VNSLVKDSTVVSERSFGMNERVRIMSWAEQMYREKEQTV 585
>ref|NP_200670.1| putative protein; protein id: At5g58620.1, supported by cDNA:
gi_15809817, supported by cDNA: gi_17064829 [Arabidopsis
thaliana] gi|8843784|dbj|BAA97332.1| zinc finger
transcription factor-like protein [Arabidopsis thaliana]
gi|15809818|gb|AAL06837.1| AT5g58620/mzn1_70
[Arabidopsis thaliana] gi|17064830|gb|AAL32569.1| zinc
finger transcription factor-like protein [Arabidopsis
thaliana] gi|21655307|gb|AAM65365.1| AT5g58620/mzn1_70
[Arabidopsis thaliana] gi|25083596|gb|AAN72094.1| zinc
finger transcription factor-like protein [Arabidopsis
thaliana]
Length = 607
Score = 113 bits (282), Expect = 5e-24
Identities = 72/155 (46%), Positives = 89/155 (56%), Gaps = 1/155 (0%)
Frame = -3
Query: 998 YPSNIPSSPVRKPSSLGFDSSSAVAAAVMNSRSAAFAK-RSQSFIDRGAAGTHHLGMSSP 822
+ +++ SSPV S DSS AV+ SR+A FAK RSQSFI+R HH +SS
Sbjct: 465 HSNSLSSSPVGANSLFSMDSS-----AVLASRAAEFAKQRSQSFIERNNGLNHHPAISSM 519
Query: 821 SNPSCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAE 642
+ +C +DW S GKLDW V GDEL KLRKS SF R GM + P + E
Sbjct: 520 TT-TC-----LNDWGSLDGKLDWSVQGDELQKLRKSTSFRLRAGGM-ESRLPNEGTGLEE 572
Query: 641 PDVSWVHSLVKDDLSKEMLPPWVEQLYIEQEQMVA 537
PDVSWV LVK+ + P W+EQ Y+E EQ VA
Sbjct: 573 PDVSWVEPLVKEPQETRLAPVWMEQSYMETEQTVA 607
>gb|AAO38462.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 749
Score = 72.8 bits (177), Expect = 8e-12
Identities = 46/116 (39%), Positives = 61/116 (51%), Gaps = 1/116 (0%)
Frame = -3
Query: 950 GFDSSSAVAAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGM-SSPSNPSCRVSSGFSDWNS 774
G +S +AAA+ RS S D G + G+ SP +SS +S W S
Sbjct: 588 GSPMNSHLAAALAQREKQQQTMRSLSSRDLGPSAARASGVVGSP------LSSSWSKWGS 641
Query: 773 PSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKD 606
PSG DWGVNG+EL KLR+S+SF R+ G +PD+SWVH+LVK+
Sbjct: 642 PSGTPDWGVNGEELGKLRRSSSFELRSGG-------------DDPDLSWVHTLVKE 684
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 897,389,451
Number of Sequences: 1393205
Number of extensions: 20909206
Number of successful extensions: 58663
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 54590
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 58397
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 57393820016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)