Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
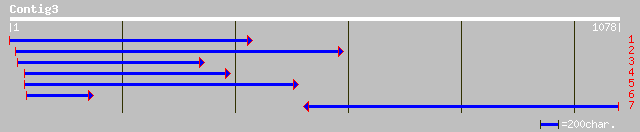
Query= KMC000602A_C03 KMC000602A_c03
(1077 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T09617 isoliquiritigenin 2'-O-methyltransferase - alfalfa g... 293 1e-83
dbj|BAA13683.1| O-methyltransferase [Glycyrrhiza echinata] 296 3e-83
pdb|1FPQ|A Chain A, Crystal Structure Analysis Of Selenomethioni... 280 8e-79
gb|AAK20170.1| caffeic acid O-methyltransferase [Catharanthus ro... 181 3e-49
emb|CAD29457.1| caffeic acid O-methyltransferase [Rosa chinensis] 178 2e-48
>pir||T09617 isoliquiritigenin 2'-O-methyltransferase - alfalfa
gi|13399462|pdb|1FP1|D Chain D, Crystal Structure
Analysis Of Chalcone O-Methyltransferase
gi|1843462|gb|AAB48059.1| isoliquiritigenin
2'-O-methyltransferase [Medicago sativa]
Length = 372
Score = 293 bits (750), Expect(2) = 1e-83
Identities = 150/263 (57%), Positives = 203/263 (77%), Gaps = 5/263 (1%)
Frame = +2
Query: 20 NSNSSEKENHVDSTNSDT-----LSAMVLGISVVFPAALNAAIELNLFDIISKGSSPQNG 184
NS ++++N + +T+ T LSAMVL ++V+PA LNAAI+LNLF+II+K + P G
Sbjct: 3 NSYITKEDNQISATSEQTEDSACLSAMVLTTNLVYPAVLNAAIDLNLFEIIAKATPP--G 60
Query: 185 GFLSASEIASELPTQHHHPDLPNRLERVLRLLASHSLLAASTRSGEDGAGSEVRVYGVSP 364
F+S SEIAS+LP H DLPNRL+R+LRLLAS+S+L ++TR+ EDG G+E RVYG+S
Sbjct: 61 AFMSPSEIASKLPASTQHSDLPNRLDRMLRLLASYSVLTSTTRTIEDG-GAE-RVYGLSM 118
Query: 365 SGQYFVSEGNRDGYLASFTSFLCHRELFRVWQNFKEAIIDPEIDLFKRVHGTSKFDHFGK 544
G+Y V + +R GYLASFT+FLC+ L +VW NFKEA++D +IDLFK VHG +K++ GK
Sbjct: 119 VGKYLVPDESR-GYLASFTTFLCYPALLQVWMNFKEAVVDEDIDLFKNVHGVTKYEFMGK 177
Query: 545 DPEMNNVSNRAMSDICSSHMNRIIEIYTGFEGISTLVDVAGGNGQGLKMIISKYPSIKGV 724
D +MN + N++M D+C++ M R++EIYTGFEGISTLVDV GG+G+ L++IISKYP IKG+
Sbjct: 178 DKKMNQIFNKSMVDVCATEMKRMLEIYTGFEGISTLVDVGGGSGRNLELIISKYPLIKGI 237
Query: 725 NFDLPQVIENSPIRPGIPSISCD 793
NFDLPQVIEN+P GI + D
Sbjct: 238 NFDLPQVIENAPPLSGIEHVGGD 260
Score = 40.0 bits (92), Expect(2) = 1e-83
Identities = 17/29 (58%), Positives = 23/29 (78%)
Frame = +1
Query: 868 GIEHVGGNMFESIPRGDAIMXKVSILNFS 954
GIEHVGG+MF S+P+GDA++ K N+S
Sbjct: 253 GIEHVGGDMFASVPQGDAMILKAVCHNWS 281
>dbj|BAA13683.1| O-methyltransferase [Glycyrrhiza echinata]
Length = 367
Score = 296 bits (758), Expect(2) = 3e-83
Identities = 153/265 (57%), Positives = 202/265 (75%), Gaps = 6/265 (2%)
Frame = +2
Query: 17 MNSNSSEKENHVDSTNSDT------LSAMVLGISVVFPAALNAAIELNLFDIISKGSSPQ 178
M+ + S K+N++ ST+S+ LSAM L ++V+PA LNAAI+LNLF+II+K + P
Sbjct: 1 MSDSYSTKDNNLFSTSSEQTEDGACLSAMRLVTNLVYPAVLNAAIDLNLFEIIAKATPP- 59
Query: 179 NGGFLSASEIASELPTQHHHPDLPNRLERVLRLLASHSLLAASTRSGEDGAGSEVRVYGV 358
G F+SASEIAS+LP H DLPNRL+R+LRLLAS+S+L +TRS E RVYG+
Sbjct: 60 -GAFMSASEIASKLPLPTQHSDLPNRLDRMLRLLASYSVLTCATRSTE-------RVYGL 111
Query: 359 SPSGQYFVSEGNRDGYLASFTSFLCHRELFRVWQNFKEAIIDPEIDLFKRVHGTSKFDHF 538
S G+Y V +G+R GYLASFT+FLC+ L VW NFKEA++D +IDLFK++HG SK+++
Sbjct: 112 SQVGKYLVPDGSR-GYLASFTTFLCYPALMNVWLNFKEAVVDEDIDLFKKLHGVSKYEYM 170
Query: 539 GKDPEMNNVSNRAMSDICSSHMNRIIEIYTGFEGISTLVDVAGGNGQGLKMIISKYPSIK 718
DP+MN++ N++M+D+C++ M RI++IY GFEGISTLVDV GGNGQ LKMIISKYP IK
Sbjct: 171 ETDPKMNHIFNKSMADVCATEMKRILQIYKGFEGISTLVDVGGGNGQNLKMIISKYPLIK 230
Query: 719 GVNFDLPQVIENSPIRPGIPSISCD 793
G+NFDLPQVIEN+P PGI + D
Sbjct: 231 GINFDLPQVIENAPPIPGIELVGGD 255
Score = 35.8 bits (81), Expect(2) = 3e-83
Identities = 16/29 (55%), Positives = 22/29 (75%)
Frame = +1
Query: 868 GIEHVGGNMFESIPRGDAIMXKVSILNFS 954
GIE VGG+MF S+P+GDA++ K N+S
Sbjct: 248 GIELVGGDMFASVPQGDAMILKAVCHNWS 276
>pdb|1FPQ|A Chain A, Crystal Structure Analysis Of Selenomethionine Substituted
Chalcone O-Methyltransferase
Length = 372
Score = 280 bits (717), Expect(2) = 8e-79
Identities = 146/263 (55%), Positives = 196/263 (74%), Gaps = 5/263 (1%)
Frame = +2
Query: 20 NSNSSEKENHVDSTNSDT-----LSAMVLGISVVFPAALNAAIELNLFDIISKGSSPQNG 184
NS ++++N + +T+ T LSA VL ++V+PA LNAAI+LNLF+II+K + P G
Sbjct: 3 NSYITKEDNQISATSEQTEDSACLSAXVLTTNLVYPAVLNAAIDLNLFEIIAKATPP--G 60
Query: 185 GFLSASEIASELPTQHHHPDLPNRLERVLRLLASHSLLAASTRSGEDGAGSEVRVYGVSP 364
F S SEIAS+LP H DLPNRL+R LRLLAS+S+L ++TR+ EDG G+E RVYG+S
Sbjct: 61 AFXSPSEIASKLPASTQHSDLPNRLDRXLRLLASYSVLTSTTRTIEDG-GAE-RVYGLSX 118
Query: 365 SGQYFVSEGNRDGYLASFTSFLCHRELFRVWQNFKEAIIDPEIDLFKRVHGTSKFDHFGK 544
G+Y V + +R GYLASFT+FLC+ L +VW NFKEA++D +IDLFK VHG +K++ GK
Sbjct: 119 VGKYLVPDESR-GYLASFTTFLCYPALLQVWXNFKEAVVDEDIDLFKNVHGVTKYEFXGK 177
Query: 545 DPEMNNVSNRAMSDICSSHMNRIIEIYTGFEGISTLVDVAGGNGQGLKMIISKYPSIKGV 724
D + N + N++ D+C++ R +EIYTGFEGISTLVDV GG+G+ L++IISKYP IKG+
Sbjct: 178 DKKXNQIFNKSXVDVCATEXKRXLEIYTGFEGISTLVDVGGGSGRNLELIISKYPLIKGI 237
Query: 725 NFDLPQVIENSPIRPGIPSISCD 793
NFDLPQVIEN+P GI + D
Sbjct: 238 NFDLPQVIENAPPLSGIEHVGGD 260
Score = 37.0 bits (84), Expect(2) = 8e-79
Identities = 16/29 (55%), Positives = 21/29 (72%)
Frame = +1
Query: 868 GIEHVGGNMFESIPRGDAIMXKVSILNFS 954
GIEHVGG+ F S+P+GDA + K N+S
Sbjct: 253 GIEHVGGDXFASVPQGDAXILKAVCHNWS 281
>gb|AAK20170.1| caffeic acid O-methyltransferase [Catharanthus roseus]
Length = 363
Score = 181 bits (460), Expect(2) = 3e-49
Identities = 107/259 (41%), Positives = 159/259 (61%), Gaps = 1/259 (0%)
Frame = +2
Query: 20 NSNSSEKENHVDSTNSDTLSAMVLGISVVFPAALNAAIELNLFDIISKGSSPQNGGFLSA 199
++N K + LSAM L + V P L +AIEL+L ++I K S P G ++S
Sbjct: 3 SANPDNKNSMTKEEEEACLSAMRLASASVLPMVLKSAIELDLLELIKK-SGP--GAYVSP 59
Query: 200 SEIASELPTQHHHPDLPNRLERVLRLLASHSLLAASTRSGEDGAGSEVRVYGVSPSGQYF 379
SE+A++LPTQ+ PD P L+R+LRLLAS+S+L + + DG G E R+Y ++P ++
Sbjct: 60 SELAAQLPTQN--PDAPVMLDRILRLLASYSVLNCTLKDLPDG-GIE-RLYSLAPVCKFL 115
Query: 380 VSEGNRDGY-LASFTSFLCHRELFRVWQNFKEAIIDPEIDLFKRVHGTSKFDHFGKDPEM 556
N DG +A+ + L W + K+A+++ I F + +G + F++ GKDP
Sbjct: 116 TK--NEDGVSMAALLLMNQDKVLMESWYHLKDAVLEGGIP-FNKAYGMTAFEYHGKDPRF 172
Query: 557 NNVSNRAMSDICSSHMNRIIEIYTGFEGISTLVDVAGGNGQGLKMIISKYPSIKGVNFDL 736
N V N+ MS+ + M +I+EIY GF+G+ T+VDV GG G L MI+SKYPSIKG+NFDL
Sbjct: 173 NKVFNQGMSNHSTIIMKKILEIYQGFQGLKTVVDVGGGTGATLNMIVSKYPSIKGINFDL 232
Query: 737 PQVIENSPIRPGIPSISCD 793
P VIE++P PG+ + D
Sbjct: 233 PHVIEDAPSYPGVDHVGGD 251
Score = 37.4 bits (85), Expect(2) = 3e-49
Identities = 14/22 (63%), Positives = 19/22 (85%)
Frame = +1
Query: 868 GIEHVGGNMFESIPRGDAIMXK 933
G++HVGG+MF S+P+GDAI K
Sbjct: 244 GVDHVGGDMFVSVPKGDAIFMK 265
>emb|CAD29457.1| caffeic acid O-methyltransferase [Rosa chinensis]
Length = 365
Score = 178 bits (452), Expect(2) = 2e-48
Identities = 103/247 (41%), Positives = 151/247 (60%), Gaps = 1/247 (0%)
Frame = +2
Query: 56 STNSDTLSAMVLGISVVFPAALNAAIELNLFDIISKGSSPQNGGFLSASEIASELPTQHH 235
S L AM L + V P L AAIEL+L +I++K G FLS +++AS+LPT++
Sbjct: 15 SDEEANLFAMQLASASVLPMVLKAAIELDLLEIMAKAGP---GAFLSPNDLASQLPTKN- 70
Query: 236 HPDLPNRLERVLRLLASHSLLAASTRSGEDGAGSEVRVYGVSPSGQYFVSEGNRDGY-LA 412
P+ P L+R+LRLLAS+S+L S R+ DG R+YG+ P ++ N DG +A
Sbjct: 71 -PEAPVMLDRMLRLLASYSILTYSLRTLPDGKVE--RLYGLGPVCKFLTK--NEDGVSIA 125
Query: 413 SFTSFLCHRELFRVWQNFKEAIIDPEIDLFKRVHGTSKFDHFGKDPEMNNVSNRAMSDIC 592
+ + L W + K+A++D I F + +G + FD+ G DP N V N+ M+D
Sbjct: 126 ALCLMNQDKVLVESWYHLKDAVLDGGIP-FNKAYGMTAFDYHGTDPRFNKVFNKGMADHS 184
Query: 593 SSHMNRIIEIYTGFEGISTLVDVAGGNGQGLKMIISKYPSIKGVNFDLPQVIENSPIRPG 772
+ M +I+E Y GFEG++++VDV GG G + MI+SKYPSIKG+NFDLP VIE++P PG
Sbjct: 185 TITMKKILETYKGFEGLTSIVDVGGGTGAVVNMIVSKYPSIKGINFDLPHVIEDAPQYPG 244
Query: 773 IPSISCD 793
+ + D
Sbjct: 245 VQHVGGD 251
Score = 37.4 bits (85), Expect(2) = 2e-48
Identities = 14/22 (63%), Positives = 19/22 (85%)
Frame = +1
Query: 868 GIEHVGGNMFESIPRGDAIMXK 933
G++HVGG+MF S+P+GDAI K
Sbjct: 244 GVQHVGGDMFVSVPKGDAIFMK 265
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 925,591,263
Number of Sequences: 1393205
Number of extensions: 20577408
Number of successful extensions: 76751
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 64291
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 76563
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 64568047518
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)