Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
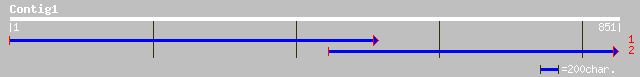
Query= KMC000561A_C01 KMC000561A_c01
(851 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC46906.1| mucin-like protein 52 9e-06
gb|EAA14411.1| agCP8278 [Anopheles gambiae str. PEST] 50 3e-05
gb|AAC02269.1| intestinal mucin [Homo sapiens] 50 3e-05
gb|AAC46904.1| mucin-like protein; Method: conceptual translatio... 50 3e-05
pir||A43932 mucin 2 precursor, intestinal - human (fragments) 49 7e-05
>gb|AAC46906.1| mucin-like protein
Length = 197
Score = 52.4 bits (124), Expect = 9e-06
Identities = 33/90 (36%), Positives = 41/90 (44%)
Frame = +3
Query: 144 PPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTTST 323
PPT TT TT P T P + + TT T + P T T TT+T
Sbjct: 78 PPTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTT-------TTTTTKPPTTTTTTTT 130
Query: 324 TPPSTTTTITTSTA*ILSS*ISSAPAIAST 413
PP+TTTT TT+TA AP+I +T
Sbjct: 131 KPPTTTTTTTTTTA-------PEAPSITTT 153
Score = 50.4 bits (119), Expect = 3e-05
Identities = 35/121 (28%), Positives = 50/121 (40%), Gaps = 2/121 (1%)
Frame = +3
Query: 63 SSSCSVNETLPSSLFSPQFRHSPLLPSPPTVTTLTT--PPSTXAGAKAPAPSLKSRXNAV 236
+ S S N T ++ P + PPT TT TT PP+T P+ +
Sbjct: 29 AESVSQNNTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTKP 88
Query: 237 RSSTTMLCIISXGTPS*MTPATCTVTTSTTPPSTTTTITTSTA*ILSS*ISSAPAIASTP 416
++TT P T T TT+T PP+TTTT TT ++ + P +T
Sbjct: 89 PTTTTTTTT---------KPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTT 139
Query: 417 T 419
T
Sbjct: 140 T 140
Score = 42.7 bits (99), Expect = 0.007
Identities = 36/142 (25%), Positives = 52/142 (36%)
Frame = +3
Query: 150 TVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTTSTTP 329
T TT TT P T P + + TT T + T T TT+T P
Sbjct: 37 TTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTT--------TTTTTTKPPTTTTTTTKP 88
Query: 330 PSTTTTITTSTA*ILSS*ISSAPAIASTPTGAGDIGVDQSRLRTPTGKPLQRAAFHFSPF 509
P+TTTT TT ++ + P +T T + + PT +P
Sbjct: 89 PTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTT--TAPE 146
Query: 510 SPV*TRRHSPHLPSPVAPDHER 575
+P T +P+ + AP R
Sbjct: 147 APSITTTEAPNTTTTRAPSSIR 168
Score = 33.5 bits (75), Expect = 4.1
Identities = 27/86 (31%), Positives = 37/86 (42%), Gaps = 2/86 (2%)
Frame = +3
Query: 144 PPTVTTLTT--PPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PPT TT TT PP+T P+ + +TT T + P ++TT
Sbjct: 99 PPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTT------TTTTTAPEAPSITT 152
Query: 318 STTPPSTTTTITTSTA*ILSS*ISSA 395
+ P +TTT +S I S SSA
Sbjct: 153 TEAPNTTTTRAPSSIRRIDGSLGSSA 178
>gb|EAA14411.1| agCP8278 [Anopheles gambiae str. PEST]
Length = 388
Score = 50.4 bits (119), Expect = 3e-05
Identities = 53/174 (30%), Positives = 77/174 (43%), Gaps = 6/174 (3%)
Frame = +3
Query: 63 SSSCSVNETLPSS---LFSPQFRHSPLLPSPPTV--TTLTTPPSTXAGAKAPAPSLKSRX 227
S + S++ T+PSS L SP R SP P+ P+ TT++T ST A S S
Sbjct: 74 SMTTSISPTIPSSPTSLSSPTTRTSPSAPTSPSTPQTTISTTESTTAVTMNSDSSSSSTE 133
Query: 228 NAVRSSTTMLCIISXGTPS*MTPATCTVTTSTTPPSTTTTITTSTA*ILSS*ISSAPAIA 407
+ SSTT + S TP + T+ TS T PS+ T T+ +A S ++ +
Sbjct: 134 STTPSSTT--------STSMTTPTSPTIPTSPTSPSSPTIRTSPSATTSPSTPKTSSSTT 185
Query: 408 STPTGAGDIGVDQSRLRTPTGKPLQRAAFHFSPFSPV*TRRHSPHLP-SPVAPD 566
+ T + + + +PT S SP T R SP P SP P+
Sbjct: 186 ESTTASSTTSISPTIPSSPT-----------SLSSP--TTRTSPSAPTSPSTPE 226
Score = 47.8 bits (112), Expect = 2e-04
Identities = 58/192 (30%), Positives = 82/192 (42%), Gaps = 24/192 (12%)
Frame = +3
Query: 63 SSSCSVNETLPSS---LFSPQFRHSPLLPSPPTV--TTLTTPPSTXAGAKAPAPSLKSRX 227
SS+ S++ T+PSS L SP R SP P+ P+ TT++T ST A + S
Sbjct: 191 SSTTSISPTIPSSPTSLSSPTTRTSPSAPTSPSTPETTISTTESTTAVTMTSDSTSSSTE 250
Query: 228 NAVRSSTT---MLCIISXGTPS*MTP-ATCTVTTSTTPPSTT----------TTITTSTA 365
+ SSTT I T MT ++ + T STT STT TT++++T+
Sbjct: 251 STTVSSTTPETTSSITESTTAVTMTSDSSSSSTESTTASSTTPATTSSTTESTTVSSTTS 310
Query: 366 *ILSS*ISSAPAIASTPTGAGDIGVDQSRLRTPTGKPLQRAAFHFSPFSPV----*TRRH 533
SS S AI T + + + T + + P SP T R
Sbjct: 311 ETTSSRTESTTAITMTSDSTSS-STESTTASSTTSTSMTTSISPTIPSSPTSLSSPTTRT 369
Query: 534 SPHLP-SPVAPD 566
SP P SP P+
Sbjct: 370 SPSAPTSPSTPE 381
Score = 46.6 bits (109), Expect = 5e-04
Identities = 41/139 (29%), Positives = 56/139 (39%), Gaps = 15/139 (10%)
Frame = +3
Query: 63 SSSCSVNETLPS-----SLFSPQFRHSPLLPSPPTVTTLTTPPSTXAGAKAPAPSLKSRX 227
SSS S T PS S+ +P P P+ P+ T+ T PS P S +
Sbjct: 127 SSSSSTESTTPSSTTSTSMTTPTSPTIPTSPTSPSSPTIRTSPSATTSPSTPKTSSSTTE 186
Query: 228 NAVRSSTTMLC-IISXGTPS*MTPATCTVTTSTTPPST---------TTTITTSTA*ILS 377
+ SSTT + I S +P T T ++ T PST +TT T T+ S
Sbjct: 187 STTASSTTSISPTIPSSPTSLSSPTTRTSPSAPTSPSTPETTISTTESTTAVTMTSDSTS 246
Query: 378 S*ISSAPAIASTPTGAGDI 434
S S ++TP I
Sbjct: 247 SSTESTTVSSTTPETTSSI 265
Score = 46.6 bits (109), Expect = 5e-04
Identities = 37/117 (31%), Positives = 59/117 (49%)
Frame = +3
Query: 63 SSSCSVNETLPSSLFSPQFRHSPLLPSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRS 242
+SS + T SS S SP +PS PT +L++P + + + +PS +
Sbjct: 180 TSSSTTESTTASSTTSI----SPTIPSSPT--SLSSPTTRTSPSAPTSPSTPETTISTTE 233
Query: 243 STTMLCIISXGTPS*MTPATCTVTTSTTPPSTTTTITTSTA*ILSS*ISSAPAIAST 413
STT + + S T S +T + T S+T P TT++IT ST + + SS+ + ST
Sbjct: 234 STTAVTMTSDSTSS----STESTTVSSTTPETTSSITESTTAVTMTSDSSSSSTEST 286
Score = 42.4 bits (98), Expect = 0.009
Identities = 37/124 (29%), Positives = 61/124 (48%), Gaps = 5/124 (4%)
Frame = +3
Query: 63 SSSCSVNETLPSSLFSPQFRHS--PLLPSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAV 236
+SS + T SS S S P +PS PT +L++P + + + +PS +
Sbjct: 57 TSSSTTESTTASSTTSTSMTTSISPTIPSSPT--SLSSPTTRTSPSAPTSPSTPQTTIST 114
Query: 237 RSSTTMLCIISXGTPS---*MTPATCTVTTSTTPPSTTTTITTSTA*ILSS*ISSAPAIA 407
STT + + S + S TP++ T T+ TTP T+ TI TS S I ++P+
Sbjct: 115 TESTTAVTMNSDSSSSSTESTTPSSTTSTSMTTP--TSPTIPTSPTSPSSPTIRTSPSAT 172
Query: 408 STPT 419
++P+
Sbjct: 173 TSPS 176
Score = 41.6 bits (96), Expect = 0.015
Identities = 49/163 (30%), Positives = 74/163 (45%), Gaps = 4/163 (2%)
Frame = +3
Query: 87 TLPSSLFSPQFRHSPLLP---SPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTML 257
T P+S SP R SP +P S P ++ TT ST A + S + SS T
Sbjct: 32 TSPTSSSSPTIRTSPSVPTSPSTPQTSSSTT-ESTTASSTTSTSMTTSISPTIPSSPT-- 88
Query: 258 CIISXGTPS*MTPATCTVTTSTTPPSTTTTITTSTA*ILSS*ISSAPAIASTPTGAGDIG 437
S +P+ T + T+ +TP +T +T ++TA ++S SS+ ++TP+
Sbjct: 89 ---SLSSPTTRTSPSAP-TSPSTPQTTISTTESTTAVTMNSDSSSSSTESTTPS-----S 139
Query: 438 VDQSRLRTPTGKPLQRAAFHFSPFSPV*TRRHSPH-LPSPVAP 563
+ + TPT + + SP SP T R SP SP P
Sbjct: 140 TTSTSMTTPTSPTIPTSP--TSPSSP--TIRTSPSATTSPSTP 178
Score = 36.6 bits (83), Expect = 0.49
Identities = 28/92 (30%), Positives = 42/92 (45%)
Frame = +3
Query: 144 PPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTTST 323
PP LTT T P S +R+S ++ S TP T + TT +
Sbjct: 12 PPLHRRLTTSMMTPTSPTIPTSPTSSSSPTIRTSPSV-------PTSPSTPQTSSSTTES 64
Query: 324 TPPSTTTTITTSTA*ILSS*ISSAPAIASTPT 419
T S+TT+ + +T+ +S I S+P S+PT
Sbjct: 65 TTASSTTSTSMTTS--ISPTIPSSPTSLSSPT 94
Score = 32.3 bits (72), Expect = 9.2
Identities = 34/110 (30%), Positives = 50/110 (44%), Gaps = 10/110 (9%)
Frame = +3
Query: 63 SSSCSVNETLPSSLF---SPQFRHSPLLPSPPTVTTLTTPPSTXA---GAKAPAPSLKSR 224
SSS S T SS + S + S + TT + ST A + + + S +S
Sbjct: 278 SSSSSTESTTASSTTPATTSSTTESTTVSSTTSETTSSRTESTTAITMTSDSTSSSTEST 337
Query: 225 XNAVRSSTTMLCIISXGTPS*MT----PATCTVTTSTTPPSTTTTITTST 362
+ +ST+M IS PS T P T T ++ T PST TI+++T
Sbjct: 338 TASSTTSTSMTTSISPTIPSSPTSLSSPTTRTSPSAPTSPSTPETISSTT 387
>gb|AAC02269.1| intestinal mucin [Homo sapiens]
Length = 589
Score = 50.4 bits (119), Expect = 3e-05
Identities = 39/128 (30%), Positives = 62/128 (47%), Gaps = 3/128 (2%)
Frame = +3
Query: 45 IVSKFFSSSCSVNETLPSSLFSPQFRHSPLLPSPPTVTTLTTPPSTXAGAKAPAPSLKSR 224
+ S SS+ + + +S + R SPLL + PT T +TP S +P ++K
Sbjct: 409 LTSSILSSTLVPSTDMITSHTTNLTRSSPLLATLPTTITRSTPTSETTYPTSPTSTVKGS 468
Query: 225 XNAVRSSTTMLCIISXGTPS*MTPATCTVTTSTTPPST---TTTITTSTA*ILSS*ISSA 395
++R ST+M +S T T ++ T + T P+T TT IT+ + SS S+
Sbjct: 469 TTSIRYSTSMTGTLSMETSLPPTSSSLPTTETATTPTTNLVTTEITSHSTPSFSS--STI 526
Query: 396 PAIASTPT 419
+ STPT
Sbjct: 527 HSTVSTPT 534
Score = 34.3 bits (77), Expect = 2.4
Identities = 29/92 (31%), Positives = 42/92 (45%), Gaps = 1/92 (1%)
Frame = +3
Query: 150 TVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTTSTTP 329
++TT+T+PP+T T++ I TP T A + TT+T P
Sbjct: 18 SMTTMTSPPTTT-----------------NCFTSLSSKILSSTPVPSTEAIISGTTNTIP 60
Query: 330 PST-TTTITTSTA*ILSS*ISSAPAIASTPTG 422
PST TT+ T A SS +S ++PTG
Sbjct: 61 PSTLVTTLPTPNA---SSMTTSETTYPNSPTG 89
>gb|AAC46904.1| mucin-like protein; Method: conceptual translation supplied by
author gi|1585304|prf||2124391B mucin-like protein
Length = 165
Score = 50.4 bits (119), Expect = 3e-05
Identities = 34/92 (36%), Positives = 44/92 (46%), Gaps = 2/92 (2%)
Frame = +3
Query: 144 PPTVTTLTT--PPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PPT TT TT PP+T P+ + +TT T + P T T TT
Sbjct: 45 PPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTT--------TTTTTKPPTTTTTT 96
Query: 318 STTPPSTTTTITTSTA*ILSS*ISSAPAIAST 413
+T PP+TTTT TT+TA AP+I +T
Sbjct: 97 TTKPPTTTTTTTTTTA-------PEAPSITTT 121
Score = 38.1 bits (87), Expect = 0.17
Identities = 32/113 (28%), Positives = 44/113 (38%), Gaps = 13/113 (11%)
Frame = +3
Query: 63 SSSCSVNETLPSSLFSPQFRHSPLLPSPPTVTTLTT--PPSTXAGAKAPAPSLKSRXNAV 236
+ S S N T ++ P + PPT TT TT PP+T P+ +
Sbjct: 29 AESVSQNNTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTK 88
Query: 237 RSSTTMLCIISXGTPS*MTPATCTVTTSTTPP-----------STTTTITTST 362
+TT T + P T T TT+TT P +TTTT S+
Sbjct: 89 PPTTTT-------TTTTKPPTTTTTTTTTTAPEAPSITTTEAPNTTTTRAPSS 134
Score = 37.0 bits (84), Expect = 0.37
Identities = 26/94 (27%), Positives = 39/94 (40%)
Frame = +3
Query: 294 PATCTVTTSTTPPSTTTTITTSTA*ILSS*ISSAPAIASTPTGAGDIGVDQSRLRTPTGK 473
P T T TT+T PP+TTTT TT ++ + P +T T + + PT
Sbjct: 45 PPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTTTTTTTKPPTTT 104
Query: 474 PLQRAAFHFSPFSPV*TRRHSPHLPSPVAPDHER 575
+P +P T +P+ + AP R
Sbjct: 105 TTTTTT--TAPEAPSITTTEAPNTTTTRAPSSIR 136
>pir||A43932 mucin 2 precursor, intestinal - human (fragments)
Length = 3020
Score = 49.3 bits (116), Expect = 7e-05
Identities = 29/75 (38%), Positives = 40/75 (52%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPPT TT T PP+T P+P + + S+TT+ P TP+ T TT
Sbjct: 1484 PSPPTTTTTTPPPTT-----TPSPPMTTPITPPASTTTL--------PPTTTPSPPTTTT 1530
Query: 318 STTPPSTTTTITTST 362
+T PP+TT + T+T
Sbjct: 1531 TTPPPTTTPSPPTTT 1545
Score = 46.2 bits (108), Expect = 6e-04
Identities = 29/94 (30%), Positives = 45/94 (47%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPP TT T PP+T PS + + ++TT TP T + +TT
Sbjct: 1454 PSPPISTTTTPPPTTTPSPPTTTPSPPTTTPSPPTTTTT-------TPPPTTTPSPPMTT 1506
Query: 318 STTPPSTTTTITTSTA*ILSS*ISSAPAIASTPT 419
TPP++TTT+ +T + ++ P +TP+
Sbjct: 1507 PITPPASTTTLPPTTTPSPPTTTTTTPPPTTTPS 1540
Score = 46.2 bits (108), Expect = 6e-04
Identities = 27/75 (36%), Positives = 36/75 (48%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPPT T +T P ST PS TT + P+ TP+ T+TT
Sbjct: 1539 PSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTT-----TPSPPTTTTPSPPTITT 1593
Query: 318 STTPPSTTTTITTST 362
+T PP+TT + T+T
Sbjct: 1594 TTPPPTTTPSPPTTT 1608
Score = 45.8 bits (107), Expect = 8e-04
Identities = 41/134 (30%), Positives = 59/134 (43%), Gaps = 6/134 (4%)
Frame = +3
Query: 93 PSSLFSPQFRHSPLLPSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISX 272
P+S SP +P P+P + TT TP T G + P + + V + T
Sbjct: 1910 PTSTQSPNGLQAPT-PTPISTTTTVTPTPTPTGTQTPTTTPITTTTTVTPTPT-----PT 1963
Query: 273 GTPS*MTPATCTVTTSTT-PPSTTTTITTSTA*ILSS*ISSAPAIASTPTGAGD-----I 434
GT TP T +TT+TT P+ T T T ST + I++ + +TPT G I
Sbjct: 1964 GT---QTPTTVLITTTTTMTPTPTPTSTKSTT---VTPITTTTTVTATPTPTGTQTPTMI 2017
Query: 435 GVDQSRLRTPTGKP 476
+ + TPT P
Sbjct: 2018 PISTTTTVTPTPTP 2031
Score = 45.4 bits (106), Expect = 0.001
Identities = 32/113 (28%), Positives = 46/113 (40%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPPT T +T P ST PS TT + P+ TP+ TT
Sbjct: 1618 PSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTT-----TPSPPTTTTPSPPITTT 1672
Query: 318 STTPPSTTTTITTSTA*ILSS*ISSAPAIASTPTGAGDIGVDQSRLRTPTGKP 476
+T PP+TT + +T + + P+ +TP+ S TP+ P
Sbjct: 1673 TTPPPTTTPSSPITTTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPPP 1725
Score = 45.4 bits (106), Expect = 0.001
Identities = 33/97 (34%), Positives = 48/97 (49%), Gaps = 4/97 (4%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAK---APAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCT 308
PSPP TT T PP+T + P+P + ++T I + TPS T +
Sbjct: 1665 PSPPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPP 1724
Query: 309 VTTSTTP-PSTTTTITTSTA*ILSS*ISSAPAIASTP 416
TT TTP P+TT + T+T L +S+P + +TP
Sbjct: 1725 PTTMTTPSPTTTPSPPTTTMTTLPPTTTSSP-LTTTP 1760
Score = 44.3 bits (103), Expect = 0.002
Identities = 26/75 (34%), Positives = 37/75 (48%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPP TT T PP+T P+P + ++T I + TP T + ++T
Sbjct: 1406 PSPPPTTTTTLPPTTT-----PSPPTTTTTTPPPTTTPSPPITTTTTPLPTTTPSPPIST 1460
Query: 318 STTPPSTTTTITTST 362
+TTPP TTT +T
Sbjct: 1461 TTTPPPTTTPSPPTT 1475
Score = 43.5 bits (101), Expect = 0.004
Identities = 28/97 (28%), Positives = 43/97 (43%), Gaps = 3/97 (3%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAG---AKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCT 308
PSPP TT T PP+T P+P + ++T + TP T +
Sbjct: 1562 PSPPPTTTTTPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTTPPPTTTPSPP 1621
Query: 309 VTTSTTPPSTTTTITTSTA*ILSS*ISSAPAIASTPT 419
TT TPP++TTT+ +T ++ P +TP+
Sbjct: 1622 TTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTPS 1658
Score = 43.1 bits (100), Expect = 0.005
Identities = 28/94 (29%), Positives = 44/94 (46%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPPT+TT T PP+T P+P + ++T +P TP T +T
Sbjct: 1586 PSPPTITTTTPPPTTT-----PSPPTTTTTTPPPTTTP--------SPPTTTPITPPTST 1632
Query: 318 STTPPSTTTTITTSTA*ILSS*ISSAPAIASTPT 419
+T PP+TT + +T + +P +TP+
Sbjct: 1633 TTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPS 1666
Score = 41.6 bits (96), Expect = 0.015
Identities = 28/78 (35%), Positives = 37/78 (46%), Gaps = 3/78 (3%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPPT TT T PP+T P+P + S+TT+ + P T TT
Sbjct: 1523 PSPPTTTTTTPPPTTT-----PSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPPPTTT 1577
Query: 318 STTPPSTT---TTITTST 362
+ P +TT TITT+T
Sbjct: 1578 PSPPTTTTPSPPTITTTT 1595
Score = 38.9 bits (89), Expect = 0.098
Identities = 27/75 (36%), Positives = 33/75 (44%)
Frame = +3
Query: 138 PSPPTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT 317
PSPPT TT T PP+T P+P + TT + TPS T T
Sbjct: 1422 PSPPTTTTTTPPPTTT-----PSPPI----------TTTTTPLPTTTPSPPISTTTTPPP 1466
Query: 318 STTPPSTTTTITTST 362
+TTP TTT + T
Sbjct: 1467 TTTPSPPTTTPSPPT 1481
Score = 37.0 bits (84), Expect = 0.37
Identities = 28/94 (29%), Positives = 40/94 (41%), Gaps = 4/94 (4%)
Frame = +3
Query: 87 TLPSSLFSPQFRHSPLLPSPPTVTTLTTPPSTXAGAKAP----APSLKSRXNAVRSSTTM 254
T PS + SP +TT TTP ST + P PS + + ++ T
Sbjct: 1687 TTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMTT 1746
Query: 255 LCIISXGTPS*MTPATCTVTTSTTPPSTTTTITT 356
L + +P TP ++T T P +TTT TT
Sbjct: 1747 LPPTTTSSPLTTTPLPPSITPPTFSPFSTTTPTT 1780
Score = 36.2 bits (82), Expect = 0.63
Identities = 33/115 (28%), Positives = 45/115 (38%), Gaps = 23/115 (20%)
Frame = +3
Query: 141 SPPTVTTLTTPPSTXAGAKAPAPSL--------------KSRXNAVRSSTTMLCIISXGT 278
+P T TT TP T G + P L ++ V TT + + T
Sbjct: 1948 TPITTTTTVTPTPTPTGTQTPTTVLITTTTTMTPTPTPTSTKSTTVTPITTTTTVTATPT 2007
Query: 279 PS*-MTPATCTVTTSTTPPSTTTTIT--------TSTA*ILSS*ISSAPAIASTP 416
P+ TP ++T+TT T T T TSTA I S+ P +STP
Sbjct: 2008 PTGTQTPTMIPISTTTTVTPTPTPTTGSTGPPTHTSTAPIAELTTSNPPPESSTP 2062
Score = 35.4 bits (80), Expect = 1.1
Identities = 39/127 (30%), Positives = 47/127 (36%), Gaps = 2/127 (1%)
Frame = +3
Query: 138 PSPPTVTTLTT--PPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTV 311
PSPP T TT PP T P + S S T + TPS T + +
Sbjct: 1657 PSPP---TTTTPSPPITTTTTPPPTTTPSSPITTTPSPPT----TTMTTPSPTTTPSSPI 1709
Query: 312 TTSTTPPSTTTTITTSTA*ILSS*ISSAPAIASTPTGAGDIGVDQSRLRTPTGKPLQRAA 491
TT+TTP STTT T S ++ +T T TP P
Sbjct: 1710 TTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMTTLPPTTTSSPLTTTPL--PPSITP 1767
Query: 492 FHFSPFS 512
FSPFS
Sbjct: 1768 PTFSPFS 1774
Score = 34.7 bits (78), Expect = 1.8
Identities = 42/142 (29%), Positives = 56/142 (38%), Gaps = 9/142 (6%)
Frame = +3
Query: 147 PTVTTLTTPPSTXAGAKAPAPSLKSRXNAVRSSTTMLCIISXGTPS*MTPATCTVTT--- 317
PT TT +T P T AP L + SST S T S +T +T ++T
Sbjct: 2029 PTPTTGSTGPPTHTST-APIAELTTSNPPPESSTPQT---SRSTSSPLTESTTLLSTLPP 2084
Query: 318 -----STTPPST-TTTITTSTA*ILSS*ISSAPAIASTPTGAGDIGVDQSRLRTPTGKPL 479
ST PPST T TTS L S P+ ++P G G PT +
Sbjct: 2085 AIEMTSTAPPSTPTAPTTTSGGHTL----SPPPSTTTSPPGTPTRGTTTGSSSAPTPSTV 2140
Query: 480 QRAAFHFSPFSPV*TRRHSPHL 545
Q S ++P T +P +
Sbjct: 2141 QTTT--TSAWTPTPTPLSTPSI 2160
Score = 32.7 bits (73), Expect = 7.0
Identities = 32/112 (28%), Positives = 48/112 (42%), Gaps = 10/112 (8%)
Frame = +3
Query: 93 PSSLFSPQFRHSPLLP------SPPTVT----TLTTPPSTXAGAKAPAPSLKSRXNAVRS 242
P++ SP +P P PPT T TTP P+P+ + + + +
Sbjct: 1653 PTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPSPT-TTPSSPITT 1711
Query: 243 STTMLCIISXGTPS*MTPATCTVTTSTTPPSTTTTITTSTA*ILSS*ISSAP 398
+TT S TPS T + +TTP TTT+TT SS +++ P
Sbjct: 1712 TTTP---SSTTTPSPPPTTMTTPSPTTTPSPPTTTMTTLPPTTTSSPLTTTP 1760
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 759,077,460
Number of Sequences: 1393205
Number of extensions: 18701899
Number of successful extensions: 101845
Number of sequences better than 10.0: 862
Number of HSP's better than 10.0 without gapping: 70813
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 92145
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 44873636157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)