Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000559A_C01 KMC000559A_c01
(485 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
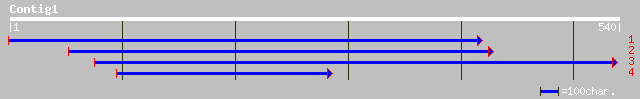
ref|NP_177694.1| leucine-rich repeat transmembrane protein kinas... 53 1e-14
ref|NP_195341.1| leucine-rich repeat transmembrane protein kinas... 44 2e-07
dbj|BAC10699.1| putative leucine rich repeat containing protein ... 43 0.002
ref|NP_190226.1| hypothetical protein; protein id: At3g46420.1 [... 42 0.005
ref|NP_181039.1| disease resistance protein family; protein id: ... 33 0.009
>ref|NP_177694.1| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At1g75640.1 [Arabidopsis thaliana]
gi|9369365|gb|AAF87114.1|AC006434_10 F10A5.16
[Arabidopsis thaliana]
Length = 1140
Score = 52.8 bits (125), Expect(2) = 1e-14
Identities = 36/81 (44%), Positives = 43/81 (52%), Gaps = 10/81 (12%)
Frame = -2
Query: 484 IPDEISKCSAWTSLVLVATE----------NLPNSTMLSLSSNSLTRKILGGFSFNSGLK 335
IPD+ISK S+ SL+L + L N T L LSSN L I S L
Sbjct: 642 IPDQISKDSSLESLLLNSNSLSGRIPESLSRLTNLTALDLSSNRLNSTIPSSLSRLRFLN 701
Query: 334 YLNLSPHNLEGEIPEMLRSRF 272
Y NLS ++LEGEIPE L +RF
Sbjct: 702 YFNLSRNSLEGEIPEALAARF 722
Score = 47.8 bits (112), Expect(2) = 1e-14
Identities = 21/39 (53%), Positives = 29/39 (73%)
Frame = -1
Query: 287 APF*IPSVFAMDERLCGKPLHIECENMRRRKRRRIIVFI 171
A F P+VF + LCGKPL IEC N+RRR+RR++I+ +
Sbjct: 720 ARFTNPTVFVKNPGLCGKPLGIECPNVRRRRRRKLILLV 758
Score = 36.2 bits (82), Expect = 0.20
Identities = 29/77 (37%), Positives = 35/77 (44%), Gaps = 10/77 (12%)
Frame = -2
Query: 484 IPDEISKCSAWTSLVL----------VATENLPNSTMLSLSSNSLTRKILGGFSFNSGLK 335
IP EI CS+ L L V L L LS NSLT I S +S L+
Sbjct: 594 IPPEIGNCSSLEVLELGSNSLKGHIPVYVSKLSLLKKLDLSHNSLTGSIPDQISKDSSLE 653
Query: 334 YLNLSPHNLEGEIPEML 284
L L+ ++L G IPE L
Sbjct: 654 SLLLNSNSLSGRIPESL 670
Score = 36.2 bits (82), Expect = 0.20
Identities = 18/40 (45%), Positives = 27/40 (67%)
Frame = -2
Query: 403 LSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIPEML 284
+ LSSN+++ KI FS +S L+ +NLS ++ GEIP L
Sbjct: 168 VDLSSNAISGKIPANFSADSSLQLINLSFNHFSGEIPATL 207
Score = 33.5 bits (75), Expect = 1.3
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = -2
Query: 421 LPNSTMLSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIPE 290
LP+ +++L +N L + GFS LKYLNLS + G IP+
Sbjct: 529 LPDLQVVALGNNLLGGVVPEGFSSLVSLKYLNLSSNLFSGHIPK 572
>ref|NP_195341.1| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At4g36180.1 [Arabidopsis thaliana]
gi|7434426|pir||T04587 hypothetical protein F23E13.70 -
Arabidopsis thaliana gi|2961377|emb|CAA18124.1| putative
receptor protein kinase [Arabidopsis thaliana]
gi|7270570|emb|CAB81527.1| putative receptor protein
kinase [Arabidopsis thaliana]
Length = 1134
Score = 43.9 bits (102), Expect(2) = 2e-07
Identities = 32/81 (39%), Positives = 43/81 (52%), Gaps = 11/81 (13%)
Frame = -2
Query: 484 IPDEISKCSAWTSLVLV----------ATENLPNSTMLSLSSNSLTRKILGGFSF-NSGL 338
IP EIS+ S+ SL L + L N T + LS N+LT +I + +S L
Sbjct: 634 IPPEISQSSSLNSLSLDHNHLSGVIPGSFSGLSNLTKMDLSVNNLTGEIPASLALISSNL 693
Query: 337 KYLNLSPHNLEGEIPEMLRSR 275
Y N+S +NL+GEIP L SR
Sbjct: 694 VYFNVSSNNLKGEIPASLGSR 714
Score = 38.9 bits (89), Expect = 0.030
Identities = 19/51 (37%), Positives = 30/51 (58%)
Frame = -2
Query: 442 VLVATENLPNSTMLSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIPE 290
V V LPN +++L N+ + + GFS L+Y+NLS ++ GEIP+
Sbjct: 514 VPVELSGLPNVQVIALQGNNFSGVVPEGFSSLVSLRYVNLSSNSFSGEIPQ 564
Score = 32.3 bits (72), Expect = 2.9
Identities = 20/47 (42%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Frame = -2
Query: 421 LPNSTM-LSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIPEML 284
LP+S L +SSN+ + +I G + + L+ LNLS + L GEIP L
Sbjct: 158 LPSSLQFLDISSNTFSGQIPSGLANLTQLQLLNLSYNQLTGEIPASL 204
Score = 31.6 bits (70), Expect(2) = 2e-07
Identities = 15/37 (40%), Positives = 25/37 (67%), Gaps = 3/37 (8%)
Frame = -1
Query: 269 SVFAMDERLCGKPLHIECENMR---RRKRRRIIVFIV 168
S F+ + LCGKPL+ CE+ ++K+R++I+ IV
Sbjct: 719 SEFSGNTELCGKPLNRRCESSTAEGKKKKRKMILMIV 755
>dbj|BAC10699.1| putative leucine rich repeat containing protein kinase [Oryza
sativa (japonica cultivar-group)]
Length = 998
Score = 42.7 bits (99), Expect = 0.002
Identities = 23/43 (53%), Positives = 30/43 (69%)
Frame = -2
Query: 421 LPNSTMLSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIP 293
+PN T + LS+N LT KI S ++GL L+LS +NLEGEIP
Sbjct: 311 MPNLTAIYLSTNELTGKIPVELSNHTGLLALDLSENNLEGEIP 353
>ref|NP_190226.1| hypothetical protein; protein id: At3g46420.1 [Arabidopsis
thaliana] gi|11346393|pir||T45699 hypothetical protein
F18L15.140 - Arabidopsis thaliana
gi|6522621|emb|CAB62033.1| hypothetical protein
[Arabidopsis thaliana]
Length = 838
Score = 41.6 bits (96), Expect = 0.005
Identities = 23/62 (37%), Positives = 35/62 (56%)
Frame = -2
Query: 460 SAWTSLVLVATENLPNSTMLSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIPEMLR 281
S T + V +NL + L LS+N+LT ++ + L ++NLS +NL G IP+ LR
Sbjct: 420 SGLTGSISVVIQNLTHLEKLDLSNNNLTGEVPDFLANMKFLVFINLSKNNLNGSIPKALR 479
Query: 280 SR 275
R
Sbjct: 480 DR 481
Score = 33.5 bits (75), Expect = 1.3
Identities = 20/49 (40%), Positives = 27/49 (54%)
Frame = -2
Query: 430 TENLPNSTMLSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIPEML 284
T P T L LSS+ LT I + L+ L+LS +NL GE+P+ L
Sbjct: 406 TSTPPRITSLDLSSSGLTGSISVVIQNLTHLEKLDLSNNNLTGEVPDFL 454
>ref|NP_181039.1| disease resistance protein family; protein id: At2g34930.1,
supported by cDNA: gi_13272422 [Arabidopsis thaliana]
gi|7485804|pir||T00475 probable disease resistance
protein [imported] - Arabidopsis thaliana
gi|3033389|gb|AAC12833.1| putative disease resistance
protein [Arabidopsis thaliana]
gi|13272423|gb|AAK17150.1|AF325082_1 putative disease
resistance protein [Arabidopsis thaliana]
gi|17380976|gb|AAL36300.1| putative disease resistance
protein [Arabidopsis thaliana]
gi|21436087|gb|AAM51244.1| putative disease resistance
protein [Arabidopsis thaliana]
Length = 905
Score = 33.1 bits (74), Expect(2) = 0.009
Identities = 18/41 (43%), Positives = 25/41 (60%)
Frame = -2
Query: 403 LSLSSNSLTRKILGGFSFNSGLKYLNLSPHNLEGEIPEMLR 281
L LS N + I F+ S L+ LNLS + LEG IP++L+
Sbjct: 838 LDLSKNKFSGAIPQSFAAISSLQRLNLSFNKLEGSIPKLLK 878
Score = 31.2 bits (69), Expect = 6.4
Identities = 22/77 (28%), Positives = 35/77 (44%), Gaps = 10/77 (12%)
Frame = -2
Query: 484 IPDEISKCSAWTSLVLVATE----------NLPNSTMLSLSSNSLTRKILGGFSFNSGLK 335
IP+ + CS T++ L + L + ML L SNS T +I L+
Sbjct: 679 IPESLRNCSGLTNIDLGGNKLTGKLPSWVGKLSSLFMLRLQSNSFTGQIPDDLCNVPNLR 738
Query: 334 YLNLSPHNLEGEIPEML 284
L+LS + + G IP+ +
Sbjct: 739 ILDLSGNKISGPIPKCI 755
Score = 26.6 bits (57), Expect(2) = 0.009
Identities = 10/19 (52%), Positives = 14/19 (73%)
Frame = -1
Query: 272 PSVFAMDERLCGKPLHIEC 216
PS++ +E LCGKPL +C
Sbjct: 882 PSIYIGNELLCGKPLPKKC 900
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 425,269,836
Number of Sequences: 1393205
Number of extensions: 9211548
Number of successful extensions: 26591
Number of sequences better than 10.0: 307
Number of HSP's better than 10.0 without gapping: 24612
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26555
length of database: 448,689,247
effective HSP length: 113
effective length of database: 291,257,082
effective search space used: 13980339936
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)