Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000551A_C01 KMC000551A_c01
(553 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
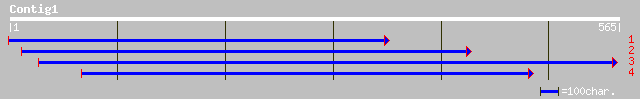
pir||C86232 hypothetical protein [imported] - Arabidopsis thalia... 42 0.004
ref|NP_704409.1| hypothetical protein [Plasmodium falciparum 3D7... 38 0.072
gb|AAL08277.1| AT5g61020/maf19_20 [Arabidopsis thaliana] 36 0.35
ref|NP_568932.1| putative protein; protein id: At5g61020.1, supp... 36 0.35
ref|NP_172452.1| unknown protein; protein id: At1g09810.1 [Arabi... 36 0.35
>pir||C86232 hypothetical protein [imported] - Arabidopsis thaliana
gi|2160172|gb|AAB60735.1| F21M12.20 gene product
[Arabidopsis thaliana]
Length = 425
Score = 42.4 bits (98), Expect = 0.004
Identities = 26/77 (33%), Positives = 38/77 (48%)
Frame = -3
Query: 548 LLDDFDFYENREKLFRSQKSTKHTSPKPQVYNSGNYHVYNNGNYHNTVEAREKKIEMQSS 369
LLDD DFYE REK R++K K + + ++ +Y GN + R +SS
Sbjct: 336 LLDDMDFYEEREKSLRAKKEHKPATLRMDLFKEKDYDYEMEGNRRMNHQERGYNWN-RSS 394
Query: 368 GGTKQFNLIKLTKNLSL 318
Q +L+ TK LS+
Sbjct: 395 NSKTQASLVNQTKYLSI 411
>ref|NP_704409.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23499148|emb|CAD51228.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 758
Score = 38.1 bits (87), Expect = 0.072
Identities = 31/98 (31%), Positives = 47/98 (47%), Gaps = 9/98 (9%)
Frame = -3
Query: 527 YENREKLFRSQKSTKHTSPKPQVYNSGN--YHVYN--NGNYHNTVEAREK----KIEMQS 372
Y+++EK R QK K K YN+ N Y YN N N HN + K K +
Sbjct: 156 YKDKEKHKREQKEKKKNKIK---YNNNNNYYFKYNSSNKNKHNYTSKKNKNNMGKKIINL 212
Query: 371 SGGTKQFNLIKLTKNLSLSPSETQLSEKQLHG-FRYGK 261
+ F +KL K+ +L+ + + EK++H FRY +
Sbjct: 213 FSLSNMFTCVKLPKDDTLTSVISSIGEKEIHSDFRYSR 250
>gb|AAL08277.1| AT5g61020/maf19_20 [Arabidopsis thaliana]
Length = 495
Score = 35.8 bits (81), Expect = 0.35
Identities = 16/29 (55%), Positives = 22/29 (75%)
Frame = -3
Query: 548 LLDDFDFYENREKLFRSQKSTKHTSPKPQ 462
+LDDF+FYENR+K+ + +KS KH K Q
Sbjct: 407 ILDDFEFYENRQKIIQERKS-KHLQIKKQ 434
>ref|NP_568932.1| putative protein; protein id: At5g61020.1, supported by cDNA:
gi_15912286 [Arabidopsis thaliana]
Length = 495
Score = 35.8 bits (81), Expect = 0.35
Identities = 16/29 (55%), Positives = 22/29 (75%)
Frame = -3
Query: 548 LLDDFDFYENREKLFRSQKSTKHTSPKPQ 462
+LDDF+FYENR+K+ + +KS KH K Q
Sbjct: 407 ILDDFEFYENRQKIIQERKS-KHLQIKKQ 434
>ref|NP_172452.1| unknown protein; protein id: At1g09810.1 [Arabidopsis thaliana]
Length = 381
Score = 35.8 bits (81), Expect = 0.35
Identities = 16/38 (42%), Positives = 23/38 (60%)
Frame = -3
Query: 548 LLDDFDFYENREKLFRSQKSTKHTSPKPQVYNSGNYHV 435
LLDD DFYE REK R++K K + + ++ +Y V
Sbjct: 339 LLDDMDFYEEREKSLRAKKEHKPATLRMDLFKEKDYDV 376
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 446,291,144
Number of Sequences: 1393205
Number of extensions: 9179672
Number of successful extensions: 27445
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 25504
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27307
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)