Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000502A_C01 KMC000502A_c01
(605 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
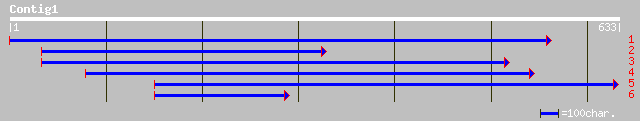
pir||T51131 ligand gated channel-like protein [imported] - rape ... 84 7e-19
pir||A84550 probable ligand-gated ion channel protein [imported]... 79 4e-17
pir||T51132 probable glutamate receptor [imported] - Arabidopsis... 79 4e-17
ref|NP_028351.1| putative ligand-gated ion channel protein; prot... 79 4e-17
ref|NP_174978.1| ligand-gated ion channel, putative; protein id:... 75 4e-16
>pir||T51131 ligand gated channel-like protein [imported] - rape
gi|6650552|gb|AAF21901.1|AF109392_1 ligand gated
channel-like protein [Brassica napus]
Length = 912
Score = 83.6 bits (205), Expect(2) = 7e-19
Identities = 42/107 (39%), Positives = 64/107 (59%), Gaps = 15/107 (14%)
Frame = -2
Query: 547 KWLSVKAC----GFRNNEDEQLQLHSFRGLFLICGITCFLALLIYFFSIVRQFSQNTPHK 380
+WLS C G ++ + EQL +HSF G+FL+CGI CF+AL I+F +VR F ++ P +
Sbjct: 799 RWLSKSNCSSPHGSQSGDSEQLNVHSFWGMFLVCGIACFVALFIHFVKVVRNFIKHKPEE 858
Query: 379 DR-----------SRIQTFLNFVDEKEDISHNQLKRKVEDISSDAYS 272
+ ++QTFL ++DEKE+ S + KRK +S +A S
Sbjct: 859 EEKDIPSPESSRLKKLQTFLAYIDEKEEESKRRFKRKRSSLSMNANS 905
Score = 32.0 bits (71), Expect(2) = 7e-19
Identities = 15/20 (75%), Positives = 17/20 (85%)
Frame = -1
Query: 605 MSTAILSLSENGELRRIRDQ 546
MSTAIL LSE GEL+RI D+
Sbjct: 780 MSTAILGLSETGELQRIHDR 799
>pir||A84550 probable ligand-gated ion channel protein [imported] - Arabidopsis
thaliana
Length = 975
Score = 78.6 bits (192), Expect(2) = 4e-17
Identities = 41/98 (41%), Positives = 59/98 (59%), Gaps = 16/98 (16%)
Frame = -2
Query: 547 KWLSVKAC----GFRNNEDEQLQLHSFRGLFLICGITCFLALLIYFFSIVRQFSQNTPH- 383
+WLS C G ++ + EQL +HSF G+FL+ GI C +AL I+FF I+R F ++TP
Sbjct: 856 RWLSKSNCSSPHGSQSGDSEQLNVHSFWGMFLVVGIACLVALFIHFFKIIRDFCKDTPEV 915
Query: 382 -----------KDRSRIQTFLNFVDEKEDISHNQLKRK 302
+++QTFL FVDEKE+ + +LKRK
Sbjct: 916 VVEEAIPSPKSSRLTKLQTFLAFVDEKEEETKRRLKRK 953
Score = 30.8 bits (68), Expect(2) = 4e-17
Identities = 14/20 (70%), Positives = 17/20 (85%)
Frame = -1
Query: 605 MSTAILSLSENGELRRIRDQ 546
MSTAIL LSE GEL++I D+
Sbjct: 837 MSTAILGLSETGELQKIHDR 856
>pir||T51132 probable glutamate receptor [imported] - Arabidopsis thaliana
gi|4185740|gb|AAD09174.1| putative glutamate receptor
[Arabidopsis thaliana]
Length = 951
Score = 78.6 bits (192), Expect(2) = 4e-17
Identities = 41/98 (41%), Positives = 59/98 (59%), Gaps = 16/98 (16%)
Frame = -2
Query: 547 KWLSVKAC----GFRNNEDEQLQLHSFRGLFLICGITCFLALLIYFFSIVRQFSQNTPH- 383
+WLS C G ++ + EQL +HSF G+FL+ GI C +AL I+FF I+R F ++TP
Sbjct: 832 RWLSKSNCSSPHGSQSGDSEQLNVHSFWGMFLVVGIACLVALFIHFFKIIRDFCKDTPEV 891
Query: 382 -----------KDRSRIQTFLNFVDEKEDISHNQLKRK 302
+++QTFL FVDEKE+ + +LKRK
Sbjct: 892 VVEEAIPSPKSSRLTKLQTFLAFVDEKEEETKRRLKRK 929
Score = 30.8 bits (68), Expect(2) = 4e-17
Identities = 14/20 (70%), Positives = 17/20 (85%)
Frame = -1
Query: 605 MSTAILSLSENGELRRIRDQ 546
MSTAIL LSE GEL++I D+
Sbjct: 813 MSTAILGLSETGELQKIHDR 832
>ref|NP_028351.1| putative ligand-gated ion channel protein; protein id: At2g17260.1,
supported by cDNA: gi_2708330 [Arabidopsis thaliana]
Length = 925
Score = 78.6 bits (192), Expect(2) = 4e-17
Identities = 41/98 (41%), Positives = 59/98 (59%), Gaps = 16/98 (16%)
Frame = -2
Query: 547 KWLSVKAC----GFRNNEDEQLQLHSFRGLFLICGITCFLALLIYFFSIVRQFSQNTPH- 383
+WLS C G ++ + EQL +HSF G+FL+ GI C +AL I+FF I+R F ++TP
Sbjct: 806 RWLSKSNCSSPHGSQSGDSEQLNVHSFWGMFLVVGIACLVALFIHFFKIIRDFCKDTPEV 865
Query: 382 -----------KDRSRIQTFLNFVDEKEDISHNQLKRK 302
+++QTFL FVDEKE+ + +LKRK
Sbjct: 866 VVEEAIPSPKSSRLTKLQTFLAFVDEKEEETKRRLKRK 903
Score = 30.8 bits (68), Expect(2) = 4e-17
Identities = 14/20 (70%), Positives = 17/20 (85%)
Frame = -1
Query: 605 MSTAILSLSENGELRRIRDQ 546
MSTAIL LSE GEL++I D+
Sbjct: 787 MSTAILGLSETGELQKIHDR 806
>ref|NP_174978.1| ligand-gated ion channel, putative; protein id: At1g42540.1
[Arabidopsis thaliana] gi|25405080|pir||C96495 probable
ligand-gated ion channel [imported] - Arabidopsis
thaliana gi|12322630|gb|AAG51316.1|AC025815_3
ligand-gated ion channel, putative [Arabidopsis thaliana]
Length = 933
Score = 75.5 bits (184), Expect(2) = 4e-16
Identities = 47/119 (39%), Positives = 60/119 (49%), Gaps = 17/119 (14%)
Frame = -2
Query: 547 KWLSVKACGFRNNEDEQLQLH--SFRGLFLICGITCFLALLIYFFSIVRQF--------- 401
KWL AC N E E +LH SF GLFLICG+ C LAL +YF I+RQ
Sbjct: 805 KWLMKNACTLENAELESDRLHLKSFWGLFLICGVACLLALFLYFVQIIRQLYKKPTDDAI 864
Query: 400 --SQNTPHKDRS----RIQTFLNFVDEKEDISHNQLKRKVEDISSDAYSRESHLRSIDQ 242
Q H S R+Q FL+ +DEKE+ H KRK++ +D S + R D+
Sbjct: 865 ARDQQQNHDSSSMRSTRLQRFLSLMDEKEESKHESKKRKIDGSMNDT-SGSTRSRGFDR 922
Score = 30.8 bits (68), Expect(2) = 4e-16
Identities = 13/20 (65%), Positives = 18/20 (90%)
Frame = -1
Query: 605 MSTAILSLSENGELRRIRDQ 546
+STAIL L+ENG+L+RI D+
Sbjct: 786 LSTAILELAENGDLQRIHDK 805
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 478,978,304
Number of Sequences: 1393205
Number of extensions: 9585000
Number of successful extensions: 25384
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 24346
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25360
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)