Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000500A_C01 KMC000500A_c01
(769 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
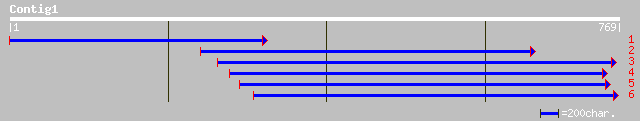
pir||T49263 hypothetical protein F12M12.190 - Arabidopsis thalia... 166 3e-40
ref|NP_566883.1| putative protein; protein id: At3g46220.1, supp... 166 3e-40
dbj|BAC38618.1| unnamed protein product [Mus musculus] 47 4e-04
dbj|BAC27976.1| unnamed protein product [Mus musculus] 47 4e-04
ref|NP_080470.1| RIKEN cDNA 1810074P20 [Mus musculus] gi|1284190... 47 4e-04
>pir||T49263 hypothetical protein F12M12.190 - Arabidopsis thaliana
gi|7799010|emb|CAB90949.1| putative protein [Arabidopsis
thaliana]
Length = 804
Score = 166 bits (420), Expect = 3e-40
Identities = 82/116 (70%), Positives = 103/116 (88%)
Frame = -1
Query: 769 HSYRKELTSQVSAETDPVSLLPKVVSLLYIQVHHKALQAPGRAISVAVSQLKDKLDESAY 590
HSYRK+L SQVS E+DP++LL KVVSLL+I++H+KALQAPGRAI+ A+S LK+KLDESAY
Sbjct: 686 HSYRKDLISQVSTESDPIALLAKVVSLLFIKIHNKALQAPGRAIAAAISHLKEKLDESAY 745
Query: 589 KILTDYQSATVTLLALLSAAPDDEESCASDRILSKRELLESKMPDLKSLVLSTSQP 422
K LTDYQ+ATVTLLAL+SA+ +E C++DRIL+KRELLES+MP L++LVL SQP
Sbjct: 746 KTLTDYQTATVTLLALMSASSGEEHDCSADRILTKRELLESQMPLLRTLVLGDSQP 801
>ref|NP_566883.1| putative protein; protein id: At3g46220.1, supported by cDNA:
gi_14334491, supported by cDNA: gi_17104790 [Arabidopsis
thaliana]
Length = 530
Score = 166 bits (420), Expect = 3e-40
Identities = 82/116 (70%), Positives = 103/116 (88%)
Frame = -1
Query: 769 HSYRKELTSQVSAETDPVSLLPKVVSLLYIQVHHKALQAPGRAISVAVSQLKDKLDESAY 590
HSYRK+L SQVS E+DP++LL KVVSLL+I++H+KALQAPGRAI+ A+S LK+KLDESAY
Sbjct: 412 HSYRKDLISQVSTESDPIALLAKVVSLLFIKIHNKALQAPGRAIAAAISHLKEKLDESAY 471
Query: 589 KILTDYQSATVTLLALLSAAPDDEESCASDRILSKRELLESKMPDLKSLVLSTSQP 422
K LTDYQ+ATVTLLAL+SA+ +E C++DRIL+KRELLES+MP L++LVL SQP
Sbjct: 472 KTLTDYQTATVTLLALMSASSGEEHDCSADRILTKRELLESQMPLLRTLVLGDSQP 527
>dbj|BAC38618.1| unnamed protein product [Mus musculus]
Length = 385
Score = 46.6 bits (109), Expect = 4e-04
Identities = 34/126 (26%), Positives = 55/126 (42%), Gaps = 9/126 (7%)
Frame = -1
Query: 763 YRKELTSQVSAETDPVSLLPKVVSLLYIQVHHKALQAPGRAISVAVSQLKDKLDESAYKI 584
+R+ L Q+ DP +L LL+ H L APGR + ++ L K+ E + +
Sbjct: 259 HRQALCEQLKVTEDPALILHLTAVLLFQLSTHSMLHAPGRCVPQIIAFLHSKIPEDQHTL 318
Query: 583 LTDYQSATVTLLALLSAAPDDEESCASDRI---------LSKRELLESKMPDLKSLVLST 431
L YQ V L + E +SD + +++EL E + +K LVL +
Sbjct: 319 LVKYQGLVVKQLVSQNKKTGQGEDPSSDELDKEQHDVTNATRKELQELSL-SIKDLVLKS 377
Query: 430 SQPS*T 413
+ S T
Sbjct: 378 RKSSVT 383
>dbj|BAC27976.1| unnamed protein product [Mus musculus]
Length = 793
Score = 46.6 bits (109), Expect = 4e-04
Identities = 34/126 (26%), Positives = 55/126 (42%), Gaps = 9/126 (7%)
Frame = -1
Query: 763 YRKELTSQVSAETDPVSLLPKVVSLLYIQVHHKALQAPGRAISVAVSQLKDKLDESAYKI 584
+R+ L Q+ DP +L LL+ H L APGR + ++ L K+ E + +
Sbjct: 667 HRQALCEQLKVTEDPALILHLTAVLLFQLSTHSMLHAPGRCVPQIIAFLHSKIPEDQHTL 726
Query: 583 LTDYQSATVTLLALLSAAPDDEESCASDRI---------LSKRELLESKMPDLKSLVLST 431
L YQ V L + E +SD + +++EL E + +K LVL +
Sbjct: 727 LVKYQGLVVKQLVSQNKKTGQGEDPSSDELDKEQHDVTNATRKELQELSL-SIKDLVLKS 785
Query: 430 SQPS*T 413
+ S T
Sbjct: 786 RKSSVT 791
>ref|NP_080470.1| RIKEN cDNA 1810074P20 [Mus musculus] gi|12841902|dbj|BAB25395.1|
unnamed protein product [Mus musculus]
Length = 793
Score = 46.6 bits (109), Expect = 4e-04
Identities = 34/126 (26%), Positives = 55/126 (42%), Gaps = 9/126 (7%)
Frame = -1
Query: 763 YRKELTSQVSAETDPVSLLPKVVSLLYIQVHHKALQAPGRAISVAVSQLKDKLDESAYKI 584
+R+ L Q+ DP +L LL+ H L APGR + ++ L K+ E + +
Sbjct: 667 HRQALCEQLKVTEDPALILHLTAVLLFQLSTHSMLHAPGRCVPQIIAFLHSKIPEDQHTL 726
Query: 583 LTDYQSATVTLLALLSAAPDDEESCASDRI---------LSKRELLESKMPDLKSLVLST 431
L YQ V L + E +SD + +++EL E + +K LVL +
Sbjct: 727 LVKYQGLVVKQLVSQNKKTGQGEDPSSDELDKEQHDVTNATRKELQELSL-SIKDLVLKS 785
Query: 430 SQPS*T 413
+ S T
Sbjct: 786 RKSSVT 791
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 631,418,245
Number of Sequences: 1393205
Number of extensions: 13224475
Number of successful extensions: 26628
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 25879
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26624
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37534933228
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)