Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000497A_C01 KMC000497A_c01
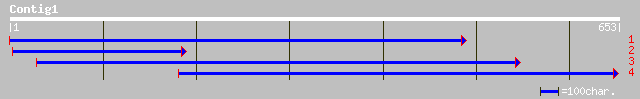
(653 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_569037.1| expressed protein; protein id: At5g66540.1, sup... 38 0.14
gb|EAA02582.1| ebiP139 [Anopheles gambiae str. PEST] 33 3.4
gb|ZP_00103302.1| hypothetical protein [Desulfitobacterium hafni... 33 4.4
ref|XP_233885.1| similar to KIAA1805 protein [Homo sapiens] [Rat... 32 5.7
gb|EAA21520.1| hypothetical protein [Plasmodium yoelii yoelii] 32 7.5
>ref|NP_569037.1| expressed protein; protein id: At5g66540.1, supported by cDNA:
gi_14488062 [Arabidopsis thaliana]
gi|10177535|dbj|BAB10930.1| gene_id:K1F13.21~unknown
protein [Arabidopsis thaliana]
gi|14488063|gb|AAK63852.1|AF389279_1 AT5g66540/K1F13_21
[Arabidopsis thaliana] gi|23506141|gb|AAN31082.1|
At5g66540/K1F13_21 [Arabidopsis thaliana]
Length = 524
Score = 37.7 bits (86), Expect = 0.14
Identities = 19/32 (59%), Positives = 24/32 (74%), Gaps = 1/32 (3%)
Frame = -2
Query: 652 DLDFEHHVRLAPVIL-EVKTSIEDMIQKMILE 560
DLDFEH+ R APVI EV S+ED+I+ I+E
Sbjct: 326 DLDFEHNARPAPVITEEVTASLEDLIKSRIIE 357
>gb|EAA02582.1| ebiP139 [Anopheles gambiae str. PEST]
Length = 422
Score = 33.1 bits (74), Expect = 3.4
Identities = 24/65 (36%), Positives = 32/65 (48%), Gaps = 3/65 (4%)
Frame = -2
Query: 235 WAIISFFHLGEAPISFMFLS*PHVLIIVSFSSVILFALFASICLITYS---MMGLGHPLY 65
WAI++ FH PI +F S + IV+ L A + + L+ S LGHPLY
Sbjct: 317 WAILAAFH---TPILHLFASKGVAIRIVTLFCCWLVASYFFVGLLFVSNTAFNNLGHPLY 373
Query: 64 SLMFN 50
S FN
Sbjct: 374 STGFN 378
>gb|ZP_00103302.1| hypothetical protein [Desulfitobacterium hafniense]
Length = 255
Score = 32.7 bits (73), Expect = 4.4
Identities = 13/26 (50%), Positives = 16/26 (61%)
Frame = +3
Query: 492 FCMELHPPPDQHLLLFPKIWHGSSRI 569
FC ELH P LL P +WH S+R+
Sbjct: 110 FCRELHVPASPSLLATPTMWHLSTRV 135
>ref|XP_233885.1| similar to KIAA1805 protein [Homo sapiens] [Rattus norvegicus]
Length = 5639
Score = 32.3 bits (72), Expect = 5.7
Identities = 24/90 (26%), Positives = 41/90 (44%), Gaps = 2/90 (2%)
Frame = +2
Query: 209 QMEKRNNSPLFMLNIHVVISQSHRPLSATEIGDNQLILLFQMIKEPAASSI*STYWLPMQ 388
Q++ SPL + Q + L+ T Q + ++ KEP A I S W+P+Q
Sbjct: 2063 QLQGEKTSPL------AIGLQEPKTLNLTSEPQPQGVTTEELNKEPQAEHIRSVQWIPLQ 2116
Query: 389 FHYLTTYLG--RPMDSQSIRR*Q*KKYLSL 472
+ +LG SQ ++R + K ++ L
Sbjct: 2117 EFHSARFLGFNSGPPSQGVKRTERKPFIKL 2146
>gb|EAA21520.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 790
Score = 32.0 bits (71), Expect = 7.5
Identities = 18/77 (23%), Positives = 38/77 (48%), Gaps = 6/77 (7%)
Frame = +1
Query: 313 INSPIPDDQRTCSKLYLIHILASHAVPLSHNIFRQTNGFSIHSKITIEKVSLSC------ 474
+N + ++ ++ YLI++L A + + F+ F+ + IT +++ L C
Sbjct: 576 VNPNVSENNHILNETYLIYLLEKTASLIIDHFFQNIFSFNFMTNITAQQLLLDCNAIENI 635
Query: 475 LYPKPSSVWNYTHLLIN 525
LY P+ + + +L IN
Sbjct: 636 LYKIPNVLTKFENLKIN 652
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 601,397,572
Number of Sequences: 1393205
Number of extensions: 13957261
Number of successful extensions: 32074
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 30967
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32067
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 28144814643
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)