Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000476A_C01 KMC000476A_c01
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
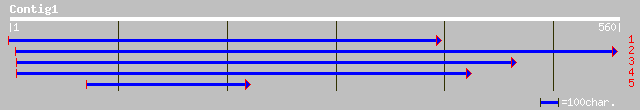
ref|NP_563958.1| expressed protein; protein id: At1g14740.1, sup... 126 2e-28
pir||F86281 protein F10B6.14 [imported] - Arabidopsis thaliana g... 126 2e-28
gb|AAO64172.1| unknown protein [Arabidopsis thaliana] 106 2e-22
ref|NP_191909.1| putative protein; protein id: At3g63500.1 [Arab... 106 2e-22
ref|NP_566320.1| expressed protein; protein id: At3g07780.1, sup... 68 7e-11
>ref|NP_563958.1| expressed protein; protein id: At1g14740.1, supported by cDNA:
gi_14596044, supported by cDNA: gi_20148718 [Arabidopsis
thaliana] gi|14596045|gb|AAK68750.1| Unknown protein
[Arabidopsis thaliana] gi|20148719|gb|AAM10250.1|
unknown protein [Arabidopsis thaliana]
Length = 733
Score = 126 bits (316), Expect = 2e-28
Identities = 63/105 (60%), Positives = 82/105 (78%)
Frame = -3
Query: 465 ENLESIVRIKEAEAKMFQTQADEARREAEGFQKMIRTKTAQMEDEYAAKLGKLCLHDTEE 286
++LES+VRIKEAE +MFQ +ADEAR EAE F++MI KT +ME+EY KL +LCL +TEE
Sbjct: 629 DSLESMVRIKEAETRMFQKKADEARIEAESFKRMIEMKTEKMEEEYTEKLARLCLQETEE 688
Query: 285 TQRKKLDEVKVLESSYVDYYKMKKRMQDEIDGLLQRMEATKQPWV 151
+R KL+E+K LE+S+ DY MK RM+ EI GLL+RME T+Q V
Sbjct: 689 RRRNKLEELKKLENSHCDYRNMKLRMEAEIAGLLKRMEVTRQQLV 733
>pir||F86281 protein F10B6.14 [imported] - Arabidopsis thaliana
gi|8778236|gb|AAF79245.1|AC006917_30 F10B6.14
[Arabidopsis thaliana]
Length = 760
Score = 126 bits (316), Expect = 2e-28
Identities = 63/105 (60%), Positives = 82/105 (78%)
Frame = -3
Query: 465 ENLESIVRIKEAEAKMFQTQADEARREAEGFQKMIRTKTAQMEDEYAAKLGKLCLHDTEE 286
++LES+VRIKEAE +MFQ +ADEAR EAE F++MI KT +ME+EY KL +LCL +TEE
Sbjct: 656 DSLESMVRIKEAETRMFQKKADEARIEAESFKRMIEMKTEKMEEEYTEKLARLCLQETEE 715
Query: 285 TQRKKLDEVKVLESSYVDYYKMKKRMQDEIDGLLQRMEATKQPWV 151
+R KL+E+K LE+S+ DY MK RM+ EI GLL+RME T+Q V
Sbjct: 716 RRRNKLEELKKLENSHCDYRNMKLRMEAEIAGLLKRMEVTRQQLV 760
>gb|AAO64172.1| unknown protein [Arabidopsis thaliana]
Length = 975
Score = 106 bits (264), Expect = 2e-22
Identities = 55/140 (39%), Positives = 92/140 (65%), Gaps = 8/140 (5%)
Frame = -3
Query: 555 HQKDLKASLLSEAKSDADIQLAAL---LRRGG-----VENLESIVRIKEAEAKMFQTQAD 400
+Q AS+ + +++ A+ L RG E LESIVR+K+AEA+MFQ +AD
Sbjct: 832 NQLKRSASVADAFHRERQVEICAVEMELERGSPKEPRFEELESIVRMKQAEAEMFQGRAD 891
Query: 399 EARREAEGFQKMIRTKTAQMEDEYAAKLGKLCLHDTEETQRKKLDEVKVLESSYVDYYKM 220
+ARREAEG +++ K ++E+EY ++GKL + D +E +R++ +E++ ++ ++Y+M
Sbjct: 892 DARREAEGLKRIAIAKKEKIEEEYNRRMGKLSMEDAQERRRRRYEELEAMQRGQREFYEM 951
Query: 219 KKRMQDEIDGLLQRMEATKQ 160
K RM++E+ GLL +ME TKQ
Sbjct: 952 KMRMEEEMRGLLTKMEMTKQ 971
>ref|NP_191909.1| putative protein; protein id: At3g63500.1 [Arabidopsis thaliana]
gi|11288851|pir||T49191 hypothetical protein MAA21.130 -
Arabidopsis thaliana gi|7573333|emb|CAB87803.1| putative
protein [Arabidopsis thaliana]
Length = 1162
Score = 106 bits (264), Expect = 2e-22
Identities = 55/140 (39%), Positives = 92/140 (65%), Gaps = 8/140 (5%)
Frame = -3
Query: 555 HQKDLKASLLSEAKSDADIQLAAL---LRRGG-----VENLESIVRIKEAEAKMFQTQAD 400
+Q AS+ + +++ A+ L RG E LESIVR+K+AEA+MFQ +AD
Sbjct: 1019 NQLKRSASVADAFHRERQVEICAVEMELERGSPKEPRFEELESIVRMKQAEAEMFQGRAD 1078
Query: 399 EARREAEGFQKMIRTKTAQMEDEYAAKLGKLCLHDTEETQRKKLDEVKVLESSYVDYYKM 220
+ARREAEG +++ K ++E+EY ++GKL + D +E +R++ +E++ ++ ++Y+M
Sbjct: 1079 DARREAEGLKRIAIAKKEKIEEEYNRRMGKLSMEDAQERRRRRYEELEAMQRGQREFYEM 1138
Query: 219 KKRMQDEIDGLLQRMEATKQ 160
K RM++E+ GLL +ME TKQ
Sbjct: 1139 KMRMEEEMRGLLTKMEMTKQ 1158
>ref|NP_566320.1| expressed protein; protein id: At3g07780.1, supported by cDNA:
gi_15028084, supported by cDNA: gi_21280842 [Arabidopsis
thaliana] gi|6466960|gb|AAF13095.1|AC009176_22 unknown
protein [Arabidopsis thaliana]
gi|6648190|gb|AAF21188.1|AC013483_12 unknown protein
[Arabidopsis thaliana] gi|15028085|gb|AAK76573.1|
unknown protein [Arabidopsis thaliana]
gi|21280843|gb|AAM44995.1| unknown protein [Arabidopsis
thaliana]
Length = 566
Score = 68.2 bits (165), Expect = 7e-11
Identities = 34/93 (36%), Positives = 61/93 (65%)
Frame = -3
Query: 522 EAKSDADIQLAALLRRGGVENLESIVRIKEAEAKMFQTQADEARREAEGFQKMIRTKTAQ 343
+AK A++Q+ ++ +E +E IVR+K+AEA+MFQ +A+EA+ EAE +++++ K +
Sbjct: 430 KAKQVAELQMERQKKKQQIEEVERIVRLKQAEAEMFQLKANEAKVEAERLERIVKAKKEK 489
Query: 342 MEDEYAAKLGKLCLHDTEETQRKKLDEVKVLES 244
E+EYA+ KL L + E + +++K ES
Sbjct: 490 TEEEYASNYLKLRLSEAEAEKEYLFEKIKEQES 522
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 507,183,457
Number of Sequences: 1393205
Number of extensions: 11554142
Number of successful extensions: 41248
Number of sequences better than 10.0: 257
Number of HSP's better than 10.0 without gapping: 39152
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 41156
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)