Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000474A_C01 KMC000474A_c01
(797 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
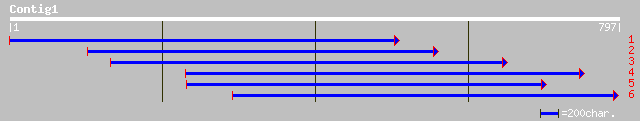
ref|NP_190169.1| DNA-binding protein - like; protein id: At3g45... 46 6e-04
ref|NP_048985.1| similar to Schizosaccharomyces ribonucleotide r... 35 1.7
gb|AAL68778.1|AF457548_1 antigen 5-related 1 protein [Anopheles ... 34 2.8
gb|EAA11396.1| agCP6145 [Anopheles gambiae str. PEST] 34 2.8
ref|NP_584637.1| hypothetical protein [Encephalitozoon cuniculi]... 34 2.8
>ref|NP_190169.1| DNA-binding protein - like; protein id: At3g45830.1 [Arabidopsis
thaliana] gi|11357267|pir||T47523 DNA-binding
protein-like - Arabidopsis thaliana
gi|7339484|emb|CAB82807.1| DNA-binding protein-like
[Arabidopsis thaliana]
Length = 1298
Score = 46.2 bits (108), Expect = 6e-04
Identities = 37/101 (36%), Positives = 52/101 (50%), Gaps = 6/101 (5%)
Frame = -1
Query: 797 AVTVAQQGTEEQXGYDLCSDLNVDPPV-IDDDKGV-EFLSSDTRPNAEDHVDVNPASEEG 624
AVTVA G EEQ ++ S+ P +D D+G + L ++T AE+ N A +G
Sbjct: 1204 AVTVAFLGNEEQTETEMGSEPKTGEPTGLDGDQGATDQLCNETEQAAEEQDGENTA--QG 1261
Query: 623 NVCEGNSMAWE---ALSLNPTATGE-LCQENSTNEDFDEES 513
N WE A+ NP +CQENS N+DFD+E+
Sbjct: 1262 N----EPTIWEPDPAVVSNPVEDNTFICQENSVNDDFDDET 1298
>ref|NP_048985.1| similar to Schizosaccharomyces ribonucleotide reductase M1 chain,
corresponds to Swiss-Prot Accession Number P36602
[Paramecium bursaria Chlorella virus 1]
gi|7461870|pir||T18131 probable
ribonucleoside-diphosphate reductase (EC 1.17.4.1) large
chain - Chlorella virus PBCV-1 gi|2447101|gb|AAC96959.1|
similar to Schizosaccharomyces ribonucleotide reductase
M1 chain, corresponds to Swiss-Prot Accession Number
P36602 [Paramecium bursaria Chlorella virus 1]
Length = 771
Score = 34.7 bits (78), Expect = 1.7
Identities = 15/43 (34%), Positives = 24/43 (54%)
Frame = -3
Query: 501 KACWPTKCKPIEKNSTGLSPLALESSSVPGSNCACI*CGRSSI 373
+ CWP KP+ K S ++ L ++ S + S CA I G S++
Sbjct: 37 RLCWPVNSKPVYKGSRAMTGLNVDVSKIVASVCASIVDGISTV 79
>gb|AAL68778.1|AF457548_1 antigen 5-related 1 protein [Anopheles gambiae]
Length = 178
Score = 33.9 bits (76), Expect = 2.8
Identities = 23/70 (32%), Positives = 31/70 (43%), Gaps = 1/70 (1%)
Frame = -1
Query: 701 GVEFLSSDTRPNAEDHVDVNPASEEGN-VCEGNSMAWEALSLNPTATGELCQENSTNEDF 525
G ++ SSD P HV NP S G C+G A + L L P + E++ N
Sbjct: 21 GGQYCSSDLCPRGGPHVGCNPPSSSGGPTCQGKQKARKVL-LTPALQAYIMDEHNLNR-- 77
Query: 524 DEESFGRERP 495
+ GR RP
Sbjct: 78 SNIALGRIRP 87
>gb|EAA11396.1| agCP6145 [Anopheles gambiae str. PEST]
Length = 260
Score = 33.9 bits (76), Expect = 2.8
Identities = 23/70 (32%), Positives = 31/70 (43%), Gaps = 1/70 (1%)
Frame = -1
Query: 701 GVEFLSSDTRPNAEDHVDVNPASEEGN-VCEGNSMAWEALSLNPTATGELCQENSTNEDF 525
G ++ SSD P HV NP S G C+G A + L L P + E++ N
Sbjct: 21 GGQYCSSDLCPRGGPHVGCNPPSSSGGPTCQGKQKARKVL-LTPALQAYIMDEHNLNR-- 77
Query: 524 DEESFGRERP 495
+ GR RP
Sbjct: 78 SNIALGRIRP 87
>ref|NP_584637.1| hypothetical protein [Encephalitozoon cuniculi]
gi|19068673|emb|CAD25141.1| hypothetical protein
[Encephalitozoon cuniculi]
Length = 603
Score = 33.9 bits (76), Expect = 2.8
Identities = 12/33 (36%), Positives = 23/33 (69%)
Frame = -1
Query: 227 SFSRSSFMWRCLLRLKILILSVYIVPL*HDDVN 129
+F R+SF+W CL+R K ++S+++ P +D +
Sbjct: 431 TFDRNSFLWSCLIRSKATLISMFLHPWVFEDTD 463
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 659,590,221
Number of Sequences: 1393205
Number of extensions: 14311004
Number of successful extensions: 34949
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 33493
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34877
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40336047648
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)