Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000430A_C01 KMC000430A_c01
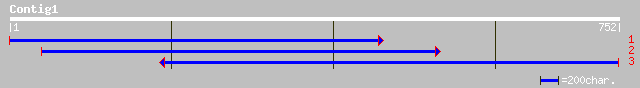
(752 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA69931.1| PR10-1 protein [Medicago truncatula] gi|13928069... 221 1e-56
sp|Q43560|PR1_MEDSA Class-10 pathogenesis-related protein 1 (MSP... 219 4e-56
pir||T09526 stress response gene 1 - alfalfa gi|1136333|gb|AAB58... 217 2e-55
emb|CAC37691.1| class 10 PR protein [Medicago sativa] 213 2e-54
gb|AAK09428.1| PR10.2C protein [Lupinus luteus] 205 5e-52
>emb|CAA69931.1| PR10-1 protein [Medicago truncatula] gi|13928069|emb|CAC37690.1|
class 10 PR protein [Medicago sativa]
Length = 157
Score = 221 bits (562), Expect = 1e-56
Identities = 107/158 (67%), Positives = 126/158 (79%)
Frame = -2
Query: 712 MAVLTFTDETTSTVAPAKLFKALVLDVDTIVPKVIPVFKSVEIVEGNGIAVGTVKKITIN 533
M V F DETTS VAPA+L+KALV D D ++PKVI +S+EIVEGNG A GT+KK+T
Sbjct: 1 MGVFNFEDETTSIVAPARLYKALVTDSDNLIPKVIDAIQSIEIVEGNGGA-GTIKKLTFV 59
Query: 532 EGGEDKYVLHKIDAIDEANFVYNYSVIGGDGLPDAAEKISFESKLIAGSDGGSIAKLTIH 353
EGGE KY LHK+D +D+ NF YNYS++GG GLPD EKISFESKL AG DGGSIAKLT+
Sbjct: 60 EGGETKYDLHKVDLVDDVNFAYNYSIVGGGGLPDTVEKISFESKLSAGPDGGSIAKLTVK 119
Query: 352 FHLKGDAKPTEAELKEGKEKGDGLFKAVEGYVLANPDY 239
+ KGDA P+E E+K GK +GDGLFKA+EGYVLANPDY
Sbjct: 120 YFTKGDAAPSEEEIKGGKARGDGLFKALEGYVLANPDY 157
>sp|Q43560|PR1_MEDSA Class-10 pathogenesis-related protein 1 (MSPR10-1)
gi|7442207|pir||T09659 pathogenesis-related protein
class 10 - alfalfa gi|1419683|emb|CAA67375.1| PR protein
from class 10 [Medicago sativa]
Length = 157
Score = 219 bits (557), Expect = 4e-56
Identities = 106/158 (67%), Positives = 125/158 (79%)
Frame = -2
Query: 712 MAVLTFTDETTSTVAPAKLFKALVLDVDTIVPKVIPVFKSVEIVEGNGIAVGTVKKITIN 533
M V F DETTS VAPA+L+KALV D D ++PKVI +S+EIVEGNG A GT+KK+T
Sbjct: 1 MGVFNFEDETTSIVAPARLYKALVTDSDNLIPKVIDAIQSIEIVEGNGGA-GTIKKLTFV 59
Query: 532 EGGEDKYVLHKIDAIDEANFVYNYSVIGGDGLPDAAEKISFESKLIAGSDGGSIAKLTIH 353
EGGE KY LHK+D +D+ NF YNYS++GG GLPD EKISFESKL AG DGGS AKLT+
Sbjct: 60 EGGETKYDLHKVDLVDDVNFAYNYSIVGGGGLPDTVEKISFESKLSAGPDGGSTAKLTVK 119
Query: 352 FHLKGDAKPTEAELKEGKEKGDGLFKAVEGYVLANPDY 239
+ KGDA P+E E+K GK +GDGLFKA+EGYVLANPDY
Sbjct: 120 YFTKGDAAPSEEEIKGGKARGDGLFKALEGYVLANPDY 157
>pir||T09526 stress response gene 1 - alfalfa gi|1136333|gb|AAB58315.1| Srg1
[Medicago sativa]
Length = 157
Score = 217 bits (552), Expect = 2e-55
Identities = 106/158 (67%), Positives = 124/158 (78%)
Frame = -2
Query: 712 MAVLTFTDETTSTVAPAKLFKALVLDVDTIVPKVIPVFKSVEIVEGNGIAVGTVKKITIN 533
M V F DETTS VAPA+L+KALV D D ++PKVI +S+EIVEGNG A GT+KK T
Sbjct: 1 MGVFNFEDETTSIVAPARLYKALVTDSDNLIPKVIDAIQSIEIVEGNGGA-GTIKKPTFV 59
Query: 532 EGGEDKYVLHKIDAIDEANFVYNYSVIGGDGLPDAAEKISFESKLIAGSDGGSIAKLTIH 353
EGGE KY LHK+D +D+ NF YNYS++GG GLPD EKISFESKL AG DGGS AKLT+
Sbjct: 60 EGGETKYDLHKVDLVDDVNFAYNYSIVGGGGLPDTVEKISFESKLSAGPDGGSTAKLTVK 119
Query: 352 FHLKGDAKPTEAELKEGKEKGDGLFKAVEGYVLANPDY 239
+ KGDA P+E E+K GK +GDGLFKA+EGYVLANPDY
Sbjct: 120 YFTKGDAAPSEEEIKGGKARGDGLFKALEGYVLANPDY 157
>emb|CAC37691.1| class 10 PR protein [Medicago sativa]
Length = 158
Score = 213 bits (542), Expect = 2e-54
Identities = 102/158 (64%), Positives = 124/158 (77%)
Frame = -2
Query: 712 MAVLTFTDETTSTVAPAKLFKALVLDVDTIVPKVIPVFKSVEIVEGNGIAVGTVKKITIN 533
M V+ F +ETTS VAPA L KA V D D ++PKV+ V KS++IVEGNG GT+KK+T
Sbjct: 1 MGVINFEEETTSIVAPATLHKAFVTDADNLIPKVVHVIKSIDIVEGNG-GSGTIKKLTFV 59
Query: 532 EGGEDKYVLHKIDAIDEANFVYNYSVIGGDGLPDAAEKISFESKLIAGSDGGSIAKLTIH 353
EGGE KY LHK+D +D+AN+ YNYS++GGD LPD EKISFE+KL AG +GGSIAKL++
Sbjct: 60 EGGETKYDLHKVDLVDDANWAYNYSIVGGDSLPDTVEKISFEAKLSAGPNGGSIAKLSVK 119
Query: 352 FHLKGDAKPTEAELKEGKEKGDGLFKAVEGYVLANPDY 239
+ KGDA P+E ELK GK KGDGLFKA+EGY LANPDY
Sbjct: 120 YFTKGDATPSEEELKSGKAKGDGLFKALEGYCLANPDY 157
>gb|AAK09428.1| PR10.2C protein [Lupinus luteus]
Length = 158
Score = 205 bits (522), Expect = 5e-52
Identities = 100/158 (63%), Positives = 121/158 (76%)
Frame = -2
Query: 712 MAVLTFTDETTSTVAPAKLFKALVLDVDTIVPKVIPVFKSVEIVEGNGIAVGTVKKITIN 533
M V TF DE+TST+APAKL+KALV D D I+PK + +SVEIVEGNG GT+KK+T
Sbjct: 1 MGVFTFQDESTSTIAPAKLYKALVTDADIIIPKAVETIQSVEIVEGNG-GPGTIKKLTFI 59
Query: 532 EGGEDKYVLHKIDAIDEANFVYNYSVIGGDGLPDAAEKISFESKLIAGSDGGSIAKLTIH 353
EGGE KYVLHKI+AIDEAN YNYS++GG GLPD EKISFE+KL+ G++GGSI K+TI
Sbjct: 60 EGGESKYVLHKIEAIDEANLGYNYSIVGGVGLPDTIEKISFETKLVEGANGGSIGKVTIK 119
Query: 352 FHLKGDAKPTEAELKEGKEKGDGLFKAVEGYVLANPDY 239
KGDA+P E E K K +GD FKA+E Y+ A+PDY
Sbjct: 120 IETKGDAQPNEEEGKAAKARGDAFFKAIESYLSAHPDY 157
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 659,171,263
Number of Sequences: 1393205
Number of extensions: 15246803
Number of successful extensions: 48681
Number of sequences better than 10.0: 308
Number of HSP's better than 10.0 without gapping: 44408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48249
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36595604110
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)