Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000428A_C02 KMC000428A_c02
(939 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
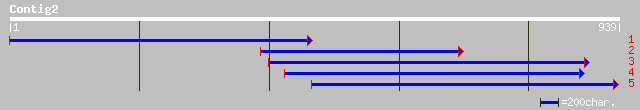
ref|NP_702384.1| hypothetical protein [Plasmodium falciparum 3D7... 35 2.2
gb|EAA16482.1| hypothetical protein [Plasmodium yoelii yoelii] 33 4.8
emb|CAD41139.1| OSJNBa0084K20.19 [Oryza sativa (japonica cultiva... 33 6.3
ref|NP_703317.1| hypothetical protein [Plasmodium falciparum 3D7... 33 6.3
>ref|NP_702384.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23497567|gb|AAN37108.1|AE014824_27 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 2189
Score = 34.7 bits (78), Expect = 2.2
Identities = 31/106 (29%), Positives = 48/106 (45%), Gaps = 8/106 (7%)
Frame = +1
Query: 55 EIERRNKYFPQSSE-------SNITSDLIFFNQ*FQNAANELNANSSTTPNEE*NSN*WT 213
E N Y P S SN S+ N N+ N N NS+T N N+N +
Sbjct: 134 ENNSNNSYQPYDSNNTNNNTNSNNNSNNNSNNNSNNNSNNYSNNNSNTNSNTNSNTN--S 191
Query: 214 *KNSISYDLHSR*KHINQRKLRRTNKILR-IRTKYDNKNKDHQSRR 348
N+ +YDL+S + I +++ +I R R + KN+D+ S +
Sbjct: 192 NNNTTNYDLNSEYEKIRRKEEEAARRIERERRADINRKNQDNNSNK 237
>gb|EAA16482.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1048
Score = 33.5 bits (75), Expect = 4.8
Identities = 32/108 (29%), Positives = 46/108 (41%), Gaps = 4/108 (3%)
Frame = +1
Query: 37 TRKKKREIERRNKYFPQ----SSESNITSDLIFFNQ*FQNAANELNANSSTTPNEE*NSN 204
++KKK+ I +N Y SS S+ SD FF N NE + NE N+N
Sbjct: 764 SKKKKKNINDQNSYISSNVSISSNSSFLSDSSFFT----NGDNENERKRKRSENESPNNN 819
Query: 205 *WT*KNSISYDLHSR*KHINQRKLRRTNKILRIRTKYDNKNKDHQSRR 348
+ KN + + K RR I + K+ NKN + Q+ R
Sbjct: 820 FYNKKNKVI-----------EIKKRRPTVIDFLNEKW-NKNSETQNVR 855
>emb|CAD41139.1| OSJNBa0084K20.19 [Oryza sativa (japonica cultivar-group)]
Length = 918
Score = 33.1 bits (74), Expect = 6.3
Identities = 21/58 (36%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Frame = -1
Query: 735 SLPLSDSPLCSLVIYPLK-CRNLR*VSLCLLDLLCLEYIAIHDFYVISCLDIGFVRLY 565
+LP S L LVI LK C NL + ++ L+ LEY+ + D Y++S + G +L+
Sbjct: 665 ALPNSIGKLSRLVILDLKACHNLEDLPKEIVKLVKLEYLDVSDCYLLSGMPKGLGKLF 722
>ref|NP_703317.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23510689|emb|CAD49074.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 637
Score = 33.1 bits (74), Expect = 6.3
Identities = 36/154 (23%), Positives = 63/154 (40%), Gaps = 14/154 (9%)
Frame = +1
Query: 43 KKKREIERRNKYFPQSSESNITSDLIFFNQ*FQNAANELNANSSTTPNEE*NSN*WT*KN 222
KKK++I NK +S +N N N+ N N+++ + N+N N
Sbjct: 153 KKKKDIIMNNKGKKNNSNNN------------NNDNNDNNDNNNSNNSNNNNNNNNNNSN 200
Query: 223 SISYDLHSR*KHINQRKLRRTNKILRIRTKYD--------------NKNKDHQSRRDAEL 360
+ ++ KH+N+ +NK KY+ NKNK+ ++ L
Sbjct: 201 EYTKRKNTHKKHLNEHYKNESNKKKVNEKKYNNSVYVNNNIKKNHVNKNKNENYLQNVWL 260
Query: 361 KLFXHCCPKYKKLKVYILGSILCLEFQNLTKKKY 462
LF + + +K +G I+ L+ N +K Y
Sbjct: 261 FLFDN---EVRKENDQCVGKIISLDTFNTIEKFY 291
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 751,838,542
Number of Sequences: 1393205
Number of extensions: 16071502
Number of successful extensions: 43307
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 40556
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43200
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 52414431048
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)