Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000422A_C01 KMC000422A_c01
(622 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
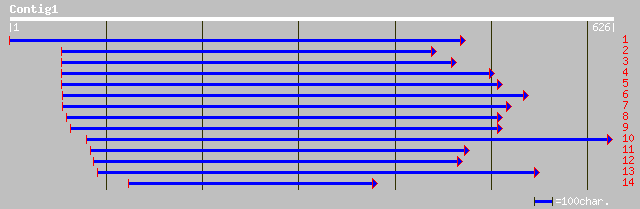
ref|NP_181616.1| glycosyl hydrolase family 77 (4-alpha-glucanotr... 129 3e-29
gb|AAL91204.1| 4-alpha-glucanotransferase [Arabidopsis thaliana] 129 3e-29
dbj|BAC22431.1| putative 4-alpha-glucanotransferase [Oryza sativ... 121 8e-27
ref|NP_811059.1| 4-alpha-glucanotransferase [Bacteroides thetaio... 68 1e-10
ref|NP_266852.1| 4-alpha-glucanotransferase (EC 2.4.1.25) [Lacto... 64 2e-09
>ref|NP_181616.1| glycosyl hydrolase family 77 (4-alpha-glucanotransferase); protein
id: At2g40840.1 [Arabidopsis thaliana]
gi|7484782|pir||T00748 4-alpha-glucanotransferase
homolog At2g40840 - Arabidopsis thaliana
gi|2623298|gb|AAB86444.1| 4-alpha-glucanotransferase
[Arabidopsis thaliana]
Length = 738
Score = 129 bits (324), Expect = 3e-29
Identities = 59/72 (81%), Positives = 65/72 (89%)
Frame = -3
Query: 620 PSMWXIFPLQDLLALKEEYTKRPAAEETINDPTNPKHYWRYRVHVTLESLLKDNELKTSI 441
PSMW IFPLQD++ALKEEYT RPA EETINDPTNPKHYWRYRVHVTL+SLLKD +LK++I
Sbjct: 642 PSMWAIFPLQDMMALKEEYTTRPATEETINDPTNPKHYWRYRVHVTLDSLLKDTDLKSTI 701
Query: 440 KNLVRWSGRSFP 405
KNLV SGRS P
Sbjct: 702 KNLVSSSGRSVP 713
>gb|AAL91204.1| 4-alpha-glucanotransferase [Arabidopsis thaliana]
Length = 955
Score = 129 bits (324), Expect = 3e-29
Identities = 59/72 (81%), Positives = 65/72 (89%)
Frame = -3
Query: 620 PSMWXIFPLQDLLALKEEYTKRPAAEETINDPTNPKHYWRYRVHVTLESLLKDNELKTSI 441
PSMW IFPLQD++ALKEEYT RPA EETINDPTNPKHYWRYRVHVTL+SLLKD +LK++I
Sbjct: 859 PSMWAIFPLQDMMALKEEYTTRPATEETINDPTNPKHYWRYRVHVTLDSLLKDTDLKSTI 918
Query: 440 KNLVRWSGRSFP 405
KNLV SGRS P
Sbjct: 919 KNLVSSSGRSVP 930
>dbj|BAC22431.1| putative 4-alpha-glucanotransferase [Oryza sativa (japonica
cultivar-group)]
Length = 922
Score = 121 bits (303), Expect = 8e-27
Identities = 56/81 (69%), Positives = 66/81 (81%)
Frame = -3
Query: 620 PSMWXIFPLQDLLALKEEYTKRPAAEETINDPTNPKHYWRYRVHVTLESLLKDNELKTSI 441
PSMW IFPLQDLLALK++YT RPA EETINDPTNPKHYWR+R+HVTL+SLL D +++ +I
Sbjct: 829 PSMWAIFPLQDLLALKDKYTTRPAKEETINDPTNPKHYWRFRLHVTLDSLLDDKDIQATI 888
Query: 440 KNLVRWSGRSFPHEDSQVEAS 378
K LV SGRSFP + E S
Sbjct: 889 KELVTSSGRSFPGKVDGAEES 909
>ref|NP_811059.1| 4-alpha-glucanotransferase [Bacteroides thetaiotaomicron VPI-5482]
gi|29339456|gb|AAO77253.1| 4-alpha-glucanotransferase
[Bacteroides thetaiotaomicron VPI-5482]
Length = 893
Score = 67.8 bits (164), Expect = 1e-10
Identities = 30/69 (43%), Positives = 47/69 (67%)
Frame = -3
Query: 617 SMWXIFPLQDLLALKEEYTKRPAAEETINDPTNPKHYWRYRVHVTLESLLKDNELKTSIK 438
S+ I LQD L++ ++ EE IN P+NP++YWRYR+H+TLE L+K EL I+
Sbjct: 822 SILCILSLQDWLSIDGKWRNPNVQEERINVPSNPRNYWRYRMHLTLEQLMKAEELNDKIR 881
Query: 437 NLVRWSGRS 411
L++++GR+
Sbjct: 882 ELIKYTGRA 890
>ref|NP_266852.1| 4-alpha-glucanotransferase (EC 2.4.1.25) [Lactococcus lactis subsp.
lactis] gi|25401203|pir||H86711
4-alpha-glucanotransferase (EC 2.4.1.25) [imported] -
Lactococcus lactis subsp. lactis (strain IL1403)
gi|12723605|gb|AAK04794.1|AE006302_12
4-alpha-glucanotransferase (EC 2.4.1.25) [Lactococcus
lactis subsp. lactis]
Length = 489
Score = 63.9 bits (154), Expect = 2e-09
Identities = 28/70 (40%), Positives = 42/70 (60%)
Frame = -3
Query: 617 SMWXIFPLQDLLALKEEYTKRPAAEETINDPTNPKHYWRYRVHVTLESLLKDNELKTSIK 438
+M I P+QD LA+ E+ K A E IN P NP HYW YR+H LE+L+ + + +K
Sbjct: 420 AMMVILPIQDWLAMSEQLRKEDAKSEQINIPANPYHYWNYRLHCQLETLIDNQDWTEFLK 479
Query: 437 NLVRWSGRSF 408
++ S R++
Sbjct: 480 KFIKESKRAY 489
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 528,119,936
Number of Sequences: 1393205
Number of extensions: 11342307
Number of successful extensions: 25348
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 24709
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25347
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)