Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000409A_C01 KMC000409A_c01
(522 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
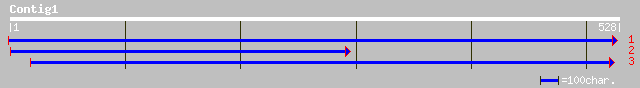
ref|NP_196024.1| expressed protein; protein id: At5g04040.1 [Ara... 48 8e-05
ref|NP_191273.1| expressed protein; protein id: At3g57140.1 [Ara... 45 5e-04
dbj|BAB61223.1| contains EST AU057376(S21389)~similar to Arabido... 39 0.028
dbj|BAB69154.1| alcC homolog (urease homolog) [Streptomyces aver... 32 4.5
ref|NP_192308.1| hypothetical protein; protein id: At4g03990.1 [... 31 7.6
>ref|NP_196024.1| expressed protein; protein id: At5g04040.1 [Arabidopsis thaliana]
gi|11282314|pir||T48431 hypothetical protein F8F6.250 -
Arabidopsis thaliana gi|7406414|emb|CAB85524.1| putative
protein [Arabidopsis thaliana]
gi|22531263|gb|AAM97135.1| putative protein [Arabidopsis
thaliana]
Length = 825
Score = 47.8 bits (112), Expect = 8e-05
Identities = 20/30 (66%), Positives = 25/30 (82%)
Frame = -2
Query: 359 SSTAKFSVSRRIPSWNCTVRENSTGSIEDL 270
+ST +F+ SRRIPSWN RENSTGS++DL
Sbjct: 595 ASTTRFNASRRIPSWNVLARENSTGSLDDL 624
>ref|NP_191273.1| expressed protein; protein id: At3g57140.1 [Arabidopsis thaliana]
gi|11282313|pir||T47774 hypothetical protein F24I3.220 -
Arabidopsis thaliana gi|6911884|emb|CAB72184.1| putative
protein [Arabidopsis thaliana]
gi|26450904|dbj|BAC42559.1| unknown protein [Arabidopsis
thaliana] gi|29029050|gb|AAO64904.1| At3g57140
[Arabidopsis thaliana]
Length = 801
Score = 45.1 bits (105), Expect = 5e-04
Identities = 17/31 (54%), Positives = 25/31 (79%)
Frame = -2
Query: 365 LKSSTAKFSVSRRIPSWNCTVRENSTGSIED 273
L + T +F+ S+RIPSWNC R+NS+GS++D
Sbjct: 597 LLAGTNRFNASKRIPSWNCIARQNSSGSVDD 627
>dbj|BAB61223.1| contains EST AU057376(S21389)~similar to Arabidopsis thaliana
chromosome 5, F8F6.250~unknown protein [Oryza sativa
(japonica cultivar-group)] gi|20804680|dbj|BAB92368.1|
P0512C01.22 [Oryza sativa (japonica cultivar-group)]
Length = 1044
Score = 39.3 bits (90), Expect = 0.028
Identities = 18/33 (54%), Positives = 23/33 (69%)
Frame = -2
Query: 353 TAKFSVSRRIPSWNCTVRENSTGSIEDLTDIAP 255
T + SRRIPSWN RENS+GS+E+ I+P
Sbjct: 592 TIRLCPSRRIPSWNLIARENSSGSLEEEFLISP 624
>dbj|BAB69154.1| alcC homolog (urease homolog) [Streptomyces avermitilis]
Length = 584
Score = 32.0 bits (71), Expect = 4.5
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 4/59 (6%)
Frame = +1
Query: 133 KILERTWKRIYLLN*LSSVPELESGVSVRNLAGV----NSSSSFSGAISVRSSMEPVEF 297
K+L W+ S VP L+ G S+ +A + ++ +SF+GA+ RS + PVE+
Sbjct: 355 KMLAALWRE-------SPVPALQEGESLATMASLVHVDHAGASFAGALIERSGLSPVEW 406
>ref|NP_192308.1| hypothetical protein; protein id: At4g03990.1 [Arabidopsis
thaliana] gi|7487394|pir||T01475 hypothetical protein
T24H24.2 - Arabidopsis thaliana
gi|3377827|gb|AAC28200.1| T24H24.2 gene product
[Arabidopsis thaliana] gi|7267155|emb|CAB77867.1|
hypothetical protein [Arabidopsis thaliana]
Length = 343
Score = 31.2 bits (69), Expect = 7.6
Identities = 15/46 (32%), Positives = 26/46 (55%)
Frame = +1
Query: 199 ESGVSVRNLAGVNSSSSFSGAISVRSSMEPVEFSLTVQFHEGILLE 336
+ G V N+ V+ SSF +VR ++EP+EF+ + G ++E
Sbjct: 14 QPGSEVDNITRVSKHSSFKIVSTVRKALEPIEFNTLMDTFLGPIVE 59
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 443,404,298
Number of Sequences: 1393205
Number of extensions: 9187979
Number of successful extensions: 22836
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 22101
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22794
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16731298976
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)