Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
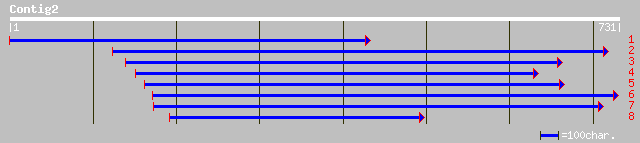
Query= KMC000408A_C02 KMC000408A_c02
(722 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM97080.1| cytoplasmic aconitate hydratase [Arabidopsis thal... 132 4e-30
ref|NP_178634.1| cytoplasmic aconitate hydratase; protein id: At... 132 4e-30
sp|P49608|ACOC_CUCMA Aconitate hydratase, cytoplasmic (Citrate h... 130 2e-29
gb|AAC26045.1| aconitase-iron regulated protein 1 [Citrus limon] 130 2e-29
gb|AAG28426.1|AF194945_1 cytosolic aconitase [Nicotiana tabacum] 128 9e-29
>gb|AAM97080.1| cytoplasmic aconitate hydratase [Arabidopsis thaliana]
Length = 990
Score = 132 bits (333), Expect = 4e-30
Identities = 62/72 (86%), Positives = 68/72 (94%)
Frame = -3
Query: 720 EDADTLGLTGHERFSIDLPNNISEIRPGQDVTVRTDSGKSFTCTVRFDTEVELEYFNHGG 541
EDADTLGLTGHER++I LP +ISEIRPGQDVTV TD+GKSFTCTVRFDTEVEL YFNHGG
Sbjct: 919 EDADTLGLTGHERYTIHLPTDISEIRPGQDVTVTTDNGKSFTCTVRFDTEVELAYFNHGG 978
Query: 540 ILPYVIRNLTRQ 505
ILPYVIRNL++Q
Sbjct: 979 ILPYVIRNLSKQ 990
>ref|NP_178634.1| cytoplasmic aconitate hydratase; protein id: At2g05710.1 [Arabidopsis
thaliana] gi|25291948|pir||B84471 cytoplasmic aconitate
hydratase [imported] - Arabidopsis thaliana
gi|4586021|gb|AAD25640.1| cytoplasmic aconitate hydratase
[Arabidopsis thaliana]
Length = 898
Score = 132 bits (333), Expect = 4e-30
Identities = 62/72 (86%), Positives = 68/72 (94%)
Frame = -3
Query: 720 EDADTLGLTGHERFSIDLPNNISEIRPGQDVTVRTDSGKSFTCTVRFDTEVELEYFNHGG 541
EDADTLGLTGHER++I LP +ISEIRPGQDVTV TD+GKSFTCTVRFDTEVEL YFNHGG
Sbjct: 827 EDADTLGLTGHERYTIHLPTDISEIRPGQDVTVTTDNGKSFTCTVRFDTEVELAYFNHGG 886
Query: 540 ILPYVIRNLTRQ 505
ILPYVIRNL++Q
Sbjct: 887 ILPYVIRNLSKQ 898
>sp|P49608|ACOC_CUCMA Aconitate hydratase, cytoplasmic (Citrate hydro-lyase) (Aconitase)
gi|7437043|pir||T10101 aconitate hydratase (EC 4.2.1.3) -
cucurbit gi|868003|dbj|BAA06108.1| aconitase [Cucurbita
cv. Kurokawa Amakuri]
Length = 898
Score = 130 bits (327), Expect = 2e-29
Identities = 61/72 (84%), Positives = 69/72 (95%)
Frame = -3
Query: 720 EDADTLGLTGHERFSIDLPNNISEIRPGQDVTVRTDSGKSFTCTVRFDTEVELEYFNHGG 541
EDAD+LGLTGHER++IDLP++IS+IRPGQDVTV TDSGKSFTCTVRFDTEVEL YFN+GG
Sbjct: 827 EDADSLGLTGHERYTIDLPDDISKIRPGQDVTVTTDSGKSFTCTVRFDTEVELAYFNNGG 886
Query: 540 ILPYVIRNLTRQ 505
ILPYVIRNL +Q
Sbjct: 887 ILPYVIRNLIKQ 898
>gb|AAC26045.1| aconitase-iron regulated protein 1 [Citrus limon]
Length = 898
Score = 130 bits (326), Expect = 2e-29
Identities = 59/72 (81%), Positives = 67/72 (92%)
Frame = -3
Query: 720 EDADTLGLTGHERFSIDLPNNISEIRPGQDVTVRTDSGKSFTCTVRFDTEVELEYFNHGG 541
EDADTLGL GHER++I+LPN +SEIRPGQD+TV TD+GKSFTCTVRFDTEVEL YF+HGG
Sbjct: 827 EDADTLGLAGHERYTINLPNKVSEIRPGQDITVTTDTGKSFTCTVRFDTEVELAYFDHGG 886
Query: 540 ILPYVIRNLTRQ 505
ILPYVIRNL +Q
Sbjct: 887 ILPYVIRNLIKQ 898
>gb|AAG28426.1|AF194945_1 cytosolic aconitase [Nicotiana tabacum]
Length = 898
Score = 128 bits (321), Expect = 9e-29
Identities = 60/72 (83%), Positives = 65/72 (89%)
Frame = -3
Query: 720 EDADTLGLTGHERFSIDLPNNISEIRPGQDVTVRTDSGKSFTCTVRFDTEVELEYFNHGG 541
EDA+TLGLTGHER++IDLP ISEI PGQDVTVRTD+GKSFTC VRFDTEVEL YFNHGG
Sbjct: 827 EDAETLGLTGHERYTIDLPEKISEIHPGQDVTVRTDTGKSFTCIVRFDTEVELAYFNHGG 886
Query: 540 ILPYVIRNLTRQ 505
ILPYVIR L +Q
Sbjct: 887 ILPYVIRQLIQQ 898
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 592,163,856
Number of Sequences: 1393205
Number of extensions: 12450501
Number of successful extensions: 30010
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 28932
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29948
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33780557640
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)