Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000400A_C02 KMC000400A_c02
(590 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
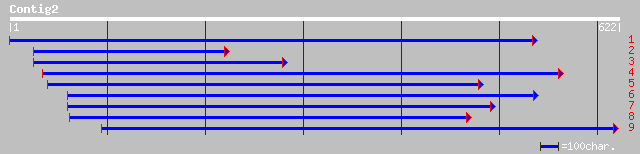
ref|NP_680446.1| Expressed protein; protein id: At5g56890.1, sup... 79 3e-14
gb|AAM19822.1| At5g56885 [Arabidopsis thaliana] gi|25090425|gb|A... 79 3e-14
dbj|BAB16871.1| putative protein kinase APK1AArabidopsis thalian... 77 1e-13
dbj|BAB39409.1| putative protein kinase [Oryza sativa (japonica ... 76 4e-13
ref|NP_179620.2| putative protein kinase; protein id: At2g20300.... 51 1e-05
>ref|NP_680446.1| Expressed protein; protein id: At5g56890.1, supported by cDNA:
gi_20453161 [Arabidopsis thaliana]
Length = 1113
Score = 79.3 bits (194), Expect = 3e-14
Identities = 53/124 (42%), Positives = 63/124 (50%), Gaps = 17/124 (13%)
Frame = -3
Query: 588 ASMCVQPEVSQRPFMGEVVQALKLVCNEFEETNYARSRR----------FQEEGLLSHVE 439
ASMCVQPEVS RPFMGEVVQALKLV NE +E S Q E
Sbjct: 976 ASMCVQPEVSHRPFMGEVVQALKLVSNECDEAKELNSLTSISKDDFRDDTQAESSCGDSS 1035
Query: 438 GKFCGY-------QSGEEKAALSPSELLSTSGQEFESFRRHSTSGPLITGKKIHFWQKLR 280
+ Y + + LS SE+ + SG+ F R S SGPL +G+ FWQK+R
Sbjct: 1036 ARMARYPLLPNYDSEPDTERGLSTSEMYTGSGR----FERQSNSGPLTSGRGKSFWQKMR 1091
Query: 279 RGST 268
R ST
Sbjct: 1092 RLST 1095
>gb|AAM19822.1| At5g56885 [Arabidopsis thaliana] gi|25090425|gb|AAN72298.1|
At5g56885/At5g56885 [Arabidopsis thaliana]
Length = 1113
Score = 79.3 bits (194), Expect = 3e-14
Identities = 53/124 (42%), Positives = 63/124 (50%), Gaps = 17/124 (13%)
Frame = -3
Query: 588 ASMCVQPEVSQRPFMGEVVQALKLVCNEFEETNYARSRR----------FQEEGLLSHVE 439
ASMCVQPEVS RPFMGEVVQALKLV NE +E S Q E
Sbjct: 976 ASMCVQPEVSHRPFMGEVVQALKLVSNECDEAKELNSLTSISKDDFRDDTQAESSCGDSS 1035
Query: 438 GKFCGY-------QSGEEKAALSPSELLSTSGQEFESFRRHSTSGPLITGKKIHFWQKLR 280
+ Y + + LS SE+ + SG+ F R S SGPL +G+ FWQK+R
Sbjct: 1036 ARMARYPLLPNYDSEPDTERGLSTSEMYTGSGR----FERQSNSGPLTSGRGKSFWQKMR 1091
Query: 279 RGST 268
R ST
Sbjct: 1092 RLST 1095
>dbj|BAB16871.1| putative protein kinase APK1AArabidopsis thaliana [Oryza sativa
(japonica cultivar-group)] gi|14646774|dbj|BAB61937.1|
putative protein kinase APK1AArabidopsis thaliana [Oryza
sativa (japonica cultivar-group)]
Length = 1014
Score = 77.4 bits (189), Expect = 1e-13
Identities = 53/114 (46%), Positives = 67/114 (58%), Gaps = 6/114 (5%)
Frame = -3
Query: 588 ASMCVQPEVSQRPFMGEVVQALKLVCNEFEETNYARSRRFQEEGLLSHVEGKFCGYQSGE 409
ASMCVQPEV+ RP MGEVVQALKLVC++ +E S F +E L + +
Sbjct: 856 ASMCVQPEVAHRPSMGEVVQALKLVCSDGDEG--LGSGSFSQE-LAAQAAAIYDVTGMEA 912
Query: 408 EKAALSP---SELLSTSGQEFESFRRHSTSGPLITGKKIHFWQKLR---RGSTS 265
E+ LS S + T + SFR+ S+SGPL+TGK FWQ+LR RGS S
Sbjct: 913 ERVLLSEMFGSTPVFTPAADSGSFRKQSSSGPLMTGKNRKFWQRLRSLSRGSMS 966
>dbj|BAB39409.1| putative protein kinase [Oryza sativa (japonica cultivar-group)]
Length = 794
Score = 75.9 bits (185), Expect = 4e-13
Identities = 52/115 (45%), Positives = 69/115 (59%), Gaps = 7/115 (6%)
Frame = -3
Query: 588 ASMCVQPEVSQRPFMGEVVQALKLVCNEFEETN----YARSRRFQEEGLLSH--VEGKFC 427
ASMCVQPEV QRPFMGEVVQALKLVC+E E N +++ Q+ G++S ++
Sbjct: 671 ASMCVQPEVDQRPFMGEVVQALKLVCDEGSEFNESGSFSQDLHIQDSGIISRASLDVDVE 730
Query: 426 GYQSGEEKAALSPSELLSTSGQEFESFRRHSTSGPLITGKKIHFWQK-LRRGSTS 265
S E A + + L SG SFRR+S+SGPL G+ H ++ L GS+S
Sbjct: 731 PVVSAELFNASAHYDTLDASG----SFRRYSSSGPLRVGRTGHNRERGLSTGSSS 781
>ref|NP_179620.2| putative protein kinase; protein id: At2g20300.1, supported by
cDNA: gi_20259542 [Arabidopsis thaliana]
gi|20259543|gb|AAM13891.1| putative protein kinase
[Arabidopsis thaliana] gi|22136896|gb|AAM91792.1|
putative protein kinase [Arabidopsis thaliana]
Length = 744
Score = 51.2 bits (121), Expect = 1e-05
Identities = 24/32 (75%), Positives = 27/32 (84%)
Frame = -3
Query: 588 ASMCVQPEVSQRPFMGEVVQALKLVCNEFEET 493
ASMCV EVS RPFMGEVVQALKL+ N+ +ET
Sbjct: 595 ASMCVHQEVSHRPFMGEVVQALKLIYNDADET 626
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 482,084,570
Number of Sequences: 1393205
Number of extensions: 9667451
Number of successful extensions: 33111
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 32612
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33102
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)