Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000397A_C01 KMC000397A_c01
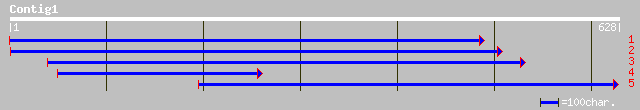
(624 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_705264.1| hypothetical protein [Plasmodium falciparum 3D7... 35 0.61
ref|NP_198672.1| putative protein; protein id: At5g38560.1, supp... 34 1.8
ref|NP_031729.1| cold inducible glycoprotein 30 [Mus musculus] g... 33 4.0
ref|XP_192861.1| elongation of very long chain fatty acids (FEN1... 33 4.0
gb|ZP_00017631.1| hypothetical protein [Chloroflexus aurantiacus] 32 6.7
>ref|NP_705264.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23615509|emb|CAD52501.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 324
Score = 35.4 bits (80), Expect = 0.61
Identities = 25/78 (32%), Positives = 37/78 (47%), Gaps = 2/78 (2%)
Frame = -3
Query: 280 CFSLFMH-FYWLHKF*NITLSKKKVLKYSKFMD*CHVYGHPFFFLA*FVL-SAFL*LAFF 107
CF+++ FYWLH + N TLSK+ LKY +Y + F + F F L F
Sbjct: 244 CFNIYNTLFYWLHIYLNFTLSKRIYLKYKT------LYKNISFVFSYFSFDEIFFFLIIF 297
Query: 106 FVKACFVSSTYQIYVLYY 53
+ F S+ + + YY
Sbjct: 298 YGFFSFTSTILTLTLFYY 315
>ref|NP_198672.1| putative protein; protein id: At5g38560.1, supported by cDNA:
gi_15983496, supported by cDNA: gi_18700152 [Arabidopsis
thaliana] gi|10176824|dbj|BAB10146.1| contains
similarity to protein kinase~gene_id:MBB18.10
[Arabidopsis thaliana]
gi|15983497|gb|AAL11616.1|AF424623_1 AT5g38560/MBB18_10
[Arabidopsis thaliana] gi|18700153|gb|AAL77688.1|
AT5g38560/MBB18_10 [Arabidopsis thaliana]
gi|21360463|gb|AAM47347.1| AT5g38560/MBB18_10
[Arabidopsis thaliana]
Length = 681
Score = 33.9 bits (76), Expect = 1.8
Identities = 18/47 (38%), Positives = 25/47 (52%)
Frame = -1
Query: 540 SQPSNRVPTVLSSPLDCGVSDHQPTSSHSPAGPILSHPSIQLRPSPP 400
S P + P V SSP VS P+SS P+ P+++ P + SPP
Sbjct: 43 SPPQSPPPVVSSSPPPPVVSSPPPSSSPPPSPPVITSPPPTVASSPP 89
>ref|NP_031729.1| cold inducible glycoprotein 30 [Mus musculus]
gi|20137691|sp|O35949|ELO3_MOUSE Elongation of very long
chain fatty acids protein 3 (Cold inducible glycoprotein
of 30 kDa) (CIN-2) gi|2289244|gb|AAC06127.1| membrane
glycoprotein CIG30 [Mus musculus]
gi|5762267|gb|AAD51088.1|AF054504_1 membrane
glycoprotein [Mus musculus] gi|16741263|gb|AAH16468.1|
elongation of very long chain fatty acids (FEN1/Elo2,
SUR4/Elo3, yeast)-like 3 [Mus musculus]
Length = 271
Score = 32.7 bits (73), Expect = 4.0
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = -1
Query: 135 LVHFSSLLFFLSKLVLYLQLTKYMFCIMRSYLYIHWIHIKSISLF 1
+V F S LF LSK+V L T ++ R +++HW H ++ LF
Sbjct: 113 VVRFWSFLFLLSKVV-ELGDTAFIILRKRPLIFVHWYHHSTVLLF 156
>ref|XP_192861.1| elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3,
yeast)-like 3 [Mus musculus]
Length = 216
Score = 32.7 bits (73), Expect = 4.0
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = -1
Query: 135 LVHFSSLLFFLSKLVLYLQLTKYMFCIMRSYLYIHWIHIKSISLF 1
+V F S LF LSK+V L T ++ R +++HW H ++ LF
Sbjct: 58 VVRFWSFLFLLSKVV-ELGDTAFIILRKRPLIFVHWYHHSTVLLF 101
>gb|ZP_00017631.1| hypothetical protein [Chloroflexus aurantiacus]
Length = 288
Score = 32.0 bits (71), Expect = 6.7
Identities = 20/52 (38%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Frame = -1
Query: 543 TSQPSNRVPTVLSSPLDCGVSDHQPTSSHSPA-GPILSHPSIQLRPSPP*GQ 391
T+QP+ +PT L SP +D TSS SPA P ++ P P+ GQ
Sbjct: 212 TAQPATAIPTALPSPTAIRQADTINTSSPSPAVTPAMTPPPAGSAPASSGGQ 263
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 536,656,091
Number of Sequences: 1393205
Number of extensions: 11806861
Number of successful extensions: 36565
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 34634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36459
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)